CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression
- PMID: 33648552
- PMCID: PMC7923339
- DOI: 10.1186/s13046-021-01884-z
CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression
Abstract
Background: Alternative polyadenylation (APA) is an important mechanism of gene expression regulation through generation of RNA isoforms with distinct 3' termini. Increasing evidence has revealed that APA is actively involved in development and disease, including hepatocellular carcinoma (HCC). However, how APA functions in tumor formation and progression remains elusive. In this study, we investigated the role of cleavage factor I (CFIm) subunit CPSF6 in human hepatocellular carcinoma (HCC).
Methods: Expression levels of CPSF6 in clinical tissues and cell lines were determined by qRT-PCR and western blot. Functional assays, including the cell number, MTT, colony formation and transwell, were used to determine the oncogenic role of CPSF6 in HCC. Animal experiments were used to determine the role of CPSF6 in HCC tumorigenicity in vivo. Deep sequencing-based 3 T-seq was used to profile the transcriptome-wide APA sites in both HCC cells and CPSF6 knockdown HCC cells. The function of CPSF6-affected target NQO1 with distinct 3'UTRs was characterized by metabolism assays.
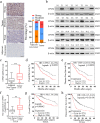
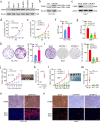
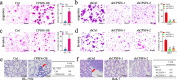
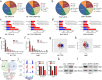
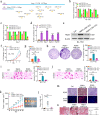
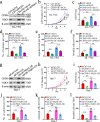
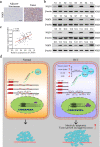
Results: We observed CPSF6 was upregulated in HCC and the high expression of CPSF6 was associated with poor prognosis in patients. Overexpression of CPSF6 promoted proliferation, migration and invasion of HCC cells in vitro and in vivo. Transcriptome-wide APA profiling analysis indicated that high expression of CPSF6 promoted the favorable usage of the proximal poly(A) site in the 3'UTR of NQO1. We demonstrated CPSF6-induced tumorigenic activities were mediated by the NQO1 isoform with short 3'UTR. Furthermore, we found that CPSF6 induced metabolic alterations in liver cells through NQO1.
Conclusion: CPSF6 plays a critical role in HCC progression by upregulating NQO1 expression through APA. These findings provide evidence to demonstrate that APA of NQO1 contributes to HCC progression and may have implications for developing new therapeutic strategy against this disease.
Keywords: Alternative polyadenylation; CPSF6; Hepatocellular carcinoma; Metabolism; NQO1.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

