MalDA, Accelerating Malaria Drug Discovery
- PMID: 33648890
- PMCID: PMC8261838
- DOI: 10.1016/j.pt.2021.01.009
MalDA, Accelerating Malaria Drug Discovery
Abstract
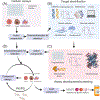
The Malaria Drug Accelerator (MalDA) is a consortium of 15 leading scientific laboratories. The aim of MalDA is to improve and accelerate the early antimalarial drug discovery process by identifying new, essential, druggable targets. In addition, it seeks to produce early lead inhibitors that may be advanced into drug candidates suitable for preclinical development and subsequent clinical testing in humans. By sharing resources, including expertise, knowledge, materials, and reagents, the consortium strives to eliminate the structural barriers often encountered in the drug discovery process. Here we discuss the mission of the consortium and its scientific achievements, including the identification of new chemically and biologically validated targets, as well as future scientific directions.
Keywords: MalDA; Plasmodium falciparum; drug resistance; malaria; target-based drug discovery.
Copyright © 2021 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Interests K.J.D. holds stock in TropIQ Health Sciences. Other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- World Health Organization (2020) World Malaria Report 2020: 20 Years of Global Progress and Challenges, WHO
-
- Paddon CJ et al. (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 - PubMed
-
- Dong Y et al. (2010) The structure−activity relationship of the antimalarial ozonide Arterolane (OZ277). J. Med. Chem. 53, 481–491 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

