Unmasking the immune microecology of ductal carcinoma in situ with deep learning
- PMID: 33649333
- PMCID: PMC7921670
- DOI: 10.1038/s41523-020-00205-5
Unmasking the immune microecology of ductal carcinoma in situ with deep learning
Abstract
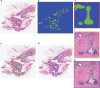
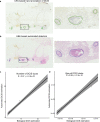
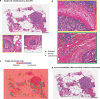
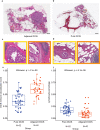
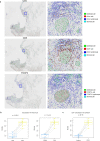
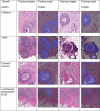
Despite increasing evidence supporting the clinical relevance of tumour infiltrating lymphocytes (TILs) in invasive breast cancer, TIL spatial variability within ductal carcinoma in situ (DCIS) samples and its association with progression are not well understood. To characterise tissue spatial architecture and the microenvironment of DCIS, we designed and validated a new deep learning pipeline, UNMaSk. Following automated detection of individual DCIS ducts using a new method IM-Net, we applied spatial tessellation to create virtual boundaries for each duct. To study local TIL infiltration for each duct, DRDIN was developed for mapping the distribution of TILs. In a dataset comprising grade 2-3 pure DCIS and DCIS adjacent to invasive cancer (adjacent DCIS), we found that pure DCIS cases had more TILs compared to adjacent DCIS. However, the colocalisation of TILs with DCIS ducts was significantly lower in pure DCIS compared to adjacent DCIS, which may suggest a more inflamed tissue ecology local to DCIS ducts in adjacent DCIS cases. Our study demonstrates that technological developments in deep convolutional neural networks and digital pathology can enable an automated morphological and microenvironmental analysis of DCIS, providing a new way to study differential immune ecology for individual ducts and identify new markers of progression.
Conflict of interest statement
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. Y.Y. has received speakers bureau honoraria from Roche and consulted for Merck and Co Inc. M.D. declares advisory fees from Radius, G1-Therapeutics, AbbVie, H3Biomedicine, Zentalis and Lilly, lecture fees from Nanostring and BCN Science and institutional grants from Pfizer and Lilly. All other authors declare that there are no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

