Human METTL7B is an alkyl thiol methyltransferase that metabolizes hydrogen sulfide and captopril
- PMID: 33649426
- PMCID: PMC7921093
- DOI: 10.1038/s41598-021-84218-5
Human METTL7B is an alkyl thiol methyltransferase that metabolizes hydrogen sulfide and captopril
Abstract
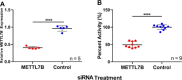
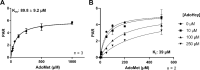
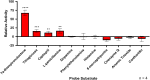
Methylation of alkyl thiols is a biotransformation pathway designed to reduce thiol reactivity and potential toxicity, yet the gene and protein responsible for human alkyl thiol methyltransferase (TMT) activity remain unknown. Here we demonstrate with a range of experimental approaches using cell lines, in vitro systems, and recombinantly expressed enzyme, that human methyltransferase-like protein 7B (METTL7B) catalyzes the transfer of a methyl group from S-adenosyl-L-methionine (AdoMet) to hydrogen sulfide (H2S) and other exogenous thiol small molecules. METTL7B gene modulation experiments, including knockdown in HepG2 cells and overexpression in HeLa cells, directly alter the methylation of the drug captopril, a historic probe substrate for TMT activity. Furthermore, recombinantly expressed and purified wild-type METTL7B methylates several thiol compounds, including H2S, 7α-thiospironolactone, L-penicillamine, and captopril, in a time- and concentration-dependent manner. Typical for AdoMet-dependent small molecule methyltransferases, S-adenosyl-L-homocysteine (AdoHcy) inhibited METTL7B activity in a competitive fashion. Similarly, mutating a conserved aspartate residue, proposed to anchor AdoMet into the active site, to an alanine (D98A) abolished methylation activity. Endogenous thiols such as glutathione and cysteine, or classic substrates for other known small molecule S-, N-, and O-methyltransferases, were not substrates for METTL7B. Our results confirm, for the first time, that METTL7B, a gene implicated in multiple disease states including rheumatoid arthritis and breast cancer, encodes a protein that methylates small molecule alkyl thiols. Identifying the catalytic function of METTL7B will enable future pharmacological research in disease pathophysiology where altered METTL7B expression and, potentially H2S levels, can disrupt cell growth and redox state.
Conflict of interest statement
The authors declare no competing interests.
Figures




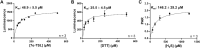
References
-
- Glauser TA, Kerremans AL, Weinshilboum RM. Human hepatic microsomal thiol methyltransferase. Assay conditions, biochemical properties, and correlation studies. Drug Metab. Dispos. 1992;20:247–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

