Handheld Point-of-Care System for Rapid Detection of SARS-CoV-2 Extracted RNA in under 20 min
- PMID: 33649735
- PMCID: PMC7839415
- DOI: 10.1021/acscentsci.0c01288
Handheld Point-of-Care System for Rapid Detection of SARS-CoV-2 Extracted RNA in under 20 min
Abstract
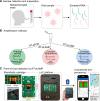
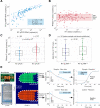
The COVID-19 pandemic is a global health emergency characterized by the high rate of transmission and ongoing increase of cases globally. Rapid point-of-care (PoC) diagnostics to detect the causative virus, SARS-CoV-2, are urgently needed to identify and isolate patients, contain its spread and guide clinical management. In this work, we report the development of a rapid PoC diagnostic test (<20 min) based on reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) and semiconductor technology for the detection of SARS-CoV-2 from extracted RNA samples. The developed LAMP assay was tested on a real-time benchtop instrument (RT-qLAMP) showing a lower limit of detection of 10 RNA copies per reaction. It was validated against extracted RNA from 183 clinical samples including 127 positive samples (screened by the CDC RT-qPCR assay). Results showed 91% sensitivity and 100% specificity when compared to RT-qPCR and average positive detection times of 15.45 ± 4.43 min. For validating the incorporation of the RT-LAMP assay onto our PoC platform (RT-eLAMP), a subset of samples was tested (n = 52), showing average detection times of 12.68 ± 2.56 min for positive samples (n = 34), demonstrating a comparable performance to a benchtop commercial instrument. Paired with a smartphone for results visualization and geolocalization, this portable diagnostic platform with secure cloud connectivity will enable real-time case identification and epidemiological surveillance.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- World Health Organization (WHO) . WHO coronavirus-2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed Dec. 28, 2020).
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

