CRISPR screens in physiologic medium reveal conditionally essential genes in human cells
- PMID: 33651980
- PMCID: PMC8172426
- DOI: 10.1016/j.cmet.2021.02.005
CRISPR screens in physiologic medium reveal conditionally essential genes in human cells
Abstract
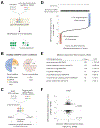
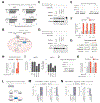
Forward genetic screens across hundreds of cancer cell lines have started to define the genetic dependencies of proliferating human cells and how these vary by genotype and lineage. Most screens, however, have been carried out in culture media that poorly reflect metabolite availability in human blood. Here, we performed CRISPR-based screens in traditional versus human plasma-like medium (HPLM). Sets of conditionally essential genes in human cancer cell lines span several cellular processes and vary with both natural cell-intrinsic diversity and the combination of basal and serum components that comprise typical media. Notably, we traced the causes for each of three conditional CRISPR phenotypes to the availability of metabolites uniquely defined in HPLM versus conventional media. Our findings reveal the profound impact of medium composition on gene essentiality in human cells, and also suggest general strategies for using genetic screens in HPLM to uncover new cancer vulnerabilities and gene-nutrient interactions.
Keywords: CRISPR; HPLM; conditional gene essentiality; gene-nutrient interaction; genetic screen; physiologic medium.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.R.C. and D.M.S. are inventors on a patent application for HPLM (PCT/US2017/061377) assigned to the Whitehead Institute. The remaining authors declare no competing interests.
Figures




References
-
- Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, et al. (2019). Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 568, 511–516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

