Characterization of a nitrite-reducing octaheme hydroxylamine oxidoreductase that lacks the tyrosine cross-link
- PMID: 33652023
- PMCID: PMC8042395
- DOI: 10.1016/j.jbc.2021.100476
Characterization of a nitrite-reducing octaheme hydroxylamine oxidoreductase that lacks the tyrosine cross-link
Abstract
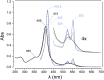
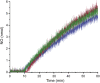
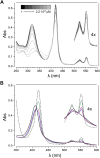
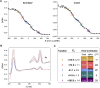
The hydroxylamine oxidoreductase (HAO) family consists of octaheme proteins that harbor seven bis-His ligated electron-transferring hemes and one 5-coordinate catalytic heme with His axial ligation. Oxidative HAOs have a homotrimeric configuration with the monomers covalently attached to each other via a unique double cross-link between a Tyr residue and the catalytic heme moiety of an adjacent subunit. This cross-linked active site heme, termed the P460 cofactor, has been hypothesized to modulate enzyme reactivity toward oxidative catalysis. Conversely, the absence of this cross-link is predicted to favor reductive catalysis. However, this prediction has not been directly tested. In this study, an HAO homolog that lacks the heme-Tyr cross-link (HAOr) was purified to homogeneity from the nitrite-dependent anaerobic ammonium-oxidizing (anammox) bacterium Kuenenia stuttgartiensis, and its catalytic and spectroscopic properties were assessed. We show that HAOr reduced nitrite to nitric oxide and also reduced nitric oxide and hydroxylamine as nonphysiological substrates. In contrast, HAOr was not able to oxidize hydroxylamine or hydrazine supporting the notion that cross-link-deficient HAO enzymes are reductases. Compared with oxidative HAOs, we found that HAOr harbors an active site heme with a higher (at least 80 mV) midpoint potential and a much lower degree of porphyrin ruffling. Based on the physiology of anammox bacteria and our results, we propose that HAOr reduces nitrite to nitric oxide in vivo, providing anammox bacteria with NO, which they use to activate ammonium in the absence of oxygen.
Keywords: HAO; anammox; cytochrome c; heme; hydroxylamine oxidoreductase; nitric oxide; nitrite reductase; nitrite reduction; redox; tyrosine cross-link.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interest.
Figures
Similar articles
-
Characterization of Anammox Hydrazine Dehydrogenase, a Key N2-producing Enzyme in the Global Nitrogen Cycle.J Biol Chem. 2016 Aug 12;291(33):17077-92. doi: 10.1074/jbc.M116.735530. Epub 2016 Jun 17. J Biol Chem. 2016. PMID: 27317665 Free PMC article.
-
Heme P460: A (Cross) Link to Nitric Oxide.Acc Chem Res. 2020 Dec 15;53(12):2925-2935. doi: 10.1021/acs.accounts.0c00573. Epub 2020 Nov 12. Acc Chem Res. 2020. PMID: 33180458 Free PMC article.
-
A 60-heme reductase complex from an anammox bacterium shows an extended electron transfer pathway.Acta Crystallogr D Struct Biol. 2019 Mar 1;75(Pt 3):333-341. doi: 10.1107/S2059798318017473. Epub 2019 Feb 28. Acta Crystallogr D Struct Biol. 2019. PMID: 30950404
-
Anammox Biochemistry: a Tale of Heme c Proteins.Trends Biochem Sci. 2016 Dec;41(12):998-1011. doi: 10.1016/j.tibs.2016.08.015. Epub 2016 Sep 23. Trends Biochem Sci. 2016. PMID: 27669648 Review.
-
Denitrification and nitrite reduction: Pseudomonas aeruginosa nitrite-reductase.Biochimie. 1984 Apr;66(4):259-89. doi: 10.1016/0300-9084(84)90005-1. Biochimie. 1984. PMID: 6331530 Review.
Cited by
-
Metagenomic evidence of a novel family of anammox bacteria in a subsea environment.Environ Microbiol. 2022 May;24(5):2348-2360. doi: 10.1111/1462-2920.16006. Epub 2022 Apr 18. Environ Microbiol. 2022. PMID: 35415863 Free PMC article.
-
Novel and unusual genes for nitrogen and metal cycling in Planctomycetota- and KSB1-affiliated metagenome-assembled genomes reconstructed from a marine subsea tunnel.FEMS Microbiol Lett. 2023 Jan 17;370:fnad049. doi: 10.1093/femsle/fnad049. FEMS Microbiol Lett. 2023. PMID: 37291701 Free PMC article.
-
Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex.Nat Microbiol. 2021 Sep;6(9):1129-1139. doi: 10.1038/s41564-021-00934-8. Epub 2021 Jul 15. Nat Microbiol. 2021. PMID: 34267357 Free PMC article.
-
Introducing Candidatus Bathyanammoxibiaceae, a family of bacteria with the anammox potential present in both marine and terrestrial environments.ISME Commun. 2022 May 19;2(1):42. doi: 10.1038/s43705-022-00125-4. ISME Commun. 2022. PMID: 37938673 Free PMC article.
-
Metagenomic Analysis of Five Phylogenetically Distant Anammox Bacterial Enrichment Cultures.Microbes Environ. 2022;37(3):ME22017. doi: 10.1264/jsme2.ME22017. Microbes Environ. 2022. PMID: 35811137 Free PMC article.
References
-
- Richardson D.J. Bacterial respiration: A flexible process for a changing environment. Microbiology. 2000;146(Pt 3):551–571. - PubMed
-
- Simon J., Kern M., Hermann B., Einsle O., Butt J.N. Physiological function and catalytic versatility of bacterial multihaem cytochromes c involved in nitrogen and sulfur cycling. Biochem. Soc. Trans. 2011;39:1864–1870. - PubMed
-
- Clarke T.A., Mills P.C., Poock S.R., Butt J.N., Cheesman M.R., Cole J.A., Hinton J.C.D., Hemmings A.M., Kemp G., Söderberg C.A.G., Spiro S., Van Wonderen J., Richardson D.J. Methods in Enzymology. Elsevier; Amsterdam, the Netherlands: 2008. Escherichia coli cytochrome c nitrite reductase NrfA; pp. 63–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

