ATF3-Induced Mammary Tumors Exhibit Molecular Features of Human Basal-Like Breast Cancer
- PMID: 33652981
- PMCID: PMC7956570
- DOI: 10.3390/ijms22052353
ATF3-Induced Mammary Tumors Exhibit Molecular Features of Human Basal-Like Breast Cancer
Abstract
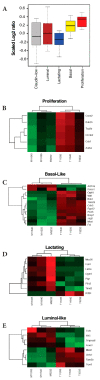
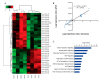
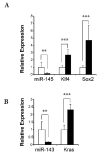
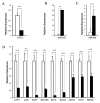
Basal-like breast cancer (BLBC) is an aggressive and deadly subtype of human breast cancer that is highly metastatic, displays stem-cell like features, and has limited treatment options. Therefore, developing and characterizing preclinical mouse models with tumors that resemble BLBC is important for human therapeutic development. ATF3 is a potent oncogene that is aberrantly expressed in most human breast cancers. In the BK5.ATF3 mouse model, overexpression of ATF3 in the basal epithelial cells of the mammary gland produces tumors that are characterized by activation of the Wnt/β-catenin signaling pathway. Here, we used RNA-Seq and microRNA (miRNA) microarrays to better define the molecular features of BK5.ATF3-derived mammary tumors. These analyses showed that these tumors share many characteristics of human BLBC including reduced expression of Rb1, Esr1, and Pgr and increased expression of Erbb2, Egfr, and the genes encoding keratins 5, 6, and 17. An analysis of miRNA expression revealed reduced levels of Mir145 and Mir143, leading to the upregulation of their target genes including both the pluripotency factors Klf4 and Sox2 as well as the cancer stem-cell-related gene Kras. Finally, we show through knock-down experiments that ATF3 may directly modulate MIR145/143 expression. Taken together, our results indicate that the ATF3 mouse mammary tumor model could provide a powerful model to define the molecular mechanisms leading to BLBC, identify the factors that contribute to its aggressiveness, and, ultimately, discover specific genes and gene networks for therapeutic targeting.
Keywords: ATF3; Mir143; Mir145; RNA-Seq; basal-like; breast cancer; miRNA; mouse model.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Badve S., Dabbs S.J., Schnitt F.L., Baehner T., Decker V., Eusebi S.B., Fox S., Ichihara J., Jacquemier S.R., Lakhani J., et al. Basal-like and triple-negative breast cancers, A critical review with an emphasis on the implications for pathologists and oncologists. Mod. Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

