Chitosan and Chitin Deacetylase Activity Are Necessary for Development and Virulence of Ustilago maydis
- PMID: 33653886
- PMCID: PMC8092297
- DOI: 10.1128/mBio.03419-20
Chitosan and Chitin Deacetylase Activity Are Necessary for Development and Virulence of Ustilago maydis
Abstract
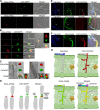
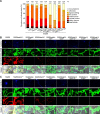
The biotrophic fungus Ustilago maydis harbors a chitin deacetylase (CDA) family of six active genes as well as one pseudogene which are differentially expressed during colonization. This includes one secreted soluble CDA (Cda4) and five putatively glycosylphosphatidylinositol (GPI)-anchored CDAs, of which Cda7 belongs to a new class of fungal CDAs. Here, we provide a comprehensive functional study of the entire family. While budding cells of U. maydis showed a discrete pattern of chitosan staining, biotrophic hyphae appeared surrounded by a chitosan layer. We purified all six active CDAs and show their activity on different chitin substrates. Single as well as multiple cda mutants were generated and revealed a virulence defect for mutants lacking cda7 We implicated cda4 in production of the chitosan layer surrounding biotrophic hyphae and demonstrated that the loss of this layer does not reduce virulence. By combining different cda mutations, we detected redundancy as well as specific functions for certain CDAs. Specifically, certain combinations of mutations significantly affected virulence concomitantly with reduced adherence, appressorium formation, penetration, and activation of plant defenses. Attempts to inactivate all seven cda genes simultaneously were unsuccessful, and induced depletion of cda2 in a background lacking the other six cda genes illustrated an essential role of chitosan for cell wall integrity.IMPORTANCE The basidiomycete Ustilago maydis causes smut disease in maize, causing substantial losses in world corn production. This nonobligate pathogen penetrates the plant cell wall with the help of appressoria and then establishes an extensive biotrophic interaction, where the hyphae are tightly encased by the plant plasma membrane. For successful invasion and development in plant tissue, recognition of conserved fungal cell wall components such as chitin by the plant immune system needs to be avoided or suppressed. One strategy to achieve this lies in the modification of chitin to chitosan by chitin deacetylases (CDAs). U. maydis has seven cda genes. This study reveals discrete as well as redundant contributions of these genes to virulence as well as to cell wall integrity. Unexpectedly, the inactivation of all seven genes is not tolerated, revealing an essential role of chitosan for viability.
Keywords: Ustilago maydis; Zea mays; chitin; chitin deacetylase; chitosan; viability; virulence.
Copyright © 2021 Rizzi et al.
Figures





References
-
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, Shimamoto K. 2013. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13:465–476. doi:10.1016/j.chom.2013.03.007. - DOI - PubMed
-
- Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K, Molinaro A, Kaku H, Shibuya N. 2014. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci U S A 111:E404–E413. doi:10.1073/pnas.1312099111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
