Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation
- PMID: 33653893
- PMCID: PMC8092269
- DOI: 10.1128/mBio.02706-20
Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation
Abstract
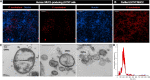
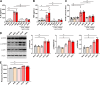
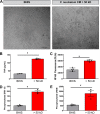
Multiple studies have implicated microbes in the development of inflammation, but the mechanisms remain unknown. Bacteria in the genus Fusobacterium have been identified in the intestinal mucosa of patients with digestive diseases; thus, we hypothesized that Fusobacterium nucleatum promotes intestinal inflammation. The addition of >50 kDa F. nucleatum conditioned media, which contain outer membrane vesicles (OMVs), to colonic epithelial cells stimulated secretion of the proinflammatory cytokines interleukin-8 (IL-8) and tumor necrosis factor (TNF). In addition, purified F. nucleatum OMVs, but not compounds <50 kDa, stimulated IL-8 and TNF production; which was decreased by pharmacological inhibition of Toll-like receptor 4 (TLR4). These effects were linked to downstream effectors p-ERK, p-CREB, and NF-κB. F. nucleatum >50-kDa compounds also stimulated TNF secretion, p-ERK, p-CREB, and NF-κB activation in human colonoid monolayers. In mice harboring a human microbiota, pretreatment with antibiotics and a single oral gavage of F. nucleatum resulted in inflammation. Compared to mice receiving vehicle control, mice treated with F. nucleatum showed disruption of the colonic architecture, with increased immune cell infiltration and depleted mucus layers. Analysis of mucosal gene expression revealed increased levels of proinflammatory cytokines (KC, TNF, IL-6, IFN-γ, and MCP-1) at day 3 and day 5 in F. nucleatum-treated mice compared to controls. These proinflammatory effects were absent in mice who received F. nucleatum without pretreatment with antibiotics, suggesting that an intact microbiome is protective against F. nucleatum-mediated immune responses. These data provide evidence that F. nucleatum promotes proinflammatory signaling cascades in the context of a depleted intestinal microbiome.IMPORTANCE Several studies have identified an increased abundance of Fusobacterium in the intestinal tracts of patients with colon cancer, liver cirrhosis, primary sclerosing cholangitis, gastroesophageal reflux disease, HIV infection, and alcoholism. However, the direct mechanism(s) of action of Fusobacterium on pathophysiological within the gastrointestinal tract is unclear. These studies have identified that F. nucleatum subsp. polymorphum releases outer membrane vesicles which activate TLR4 and NF-κB to stimulate proinflammatory signals in vitro Using mice harboring a human microbiome, we demonstrate that F. nucleatum can promote inflammation, an effect which required antibiotic-mediated alterations in the gut microbiome. Collectively, these results suggest a mechanism by which F. nucleatum may contribute to intestinal inflammation.
Keywords: Fusobacterium nucleatum; TLR4; enteroids; epithelium; inflammation; intestine; microbiome; organoids; outer membrane vesicles.
Copyright © 2021 Engevik et al.
Figures



References
-
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, Thaiss CA, Sato M, Toyooka K, Said HS, Yamagami H, Rice SA, Gevers D, Johnson RC, Segre JA, Chen K, Kolls JK, Elinav E, Morita H, Xavier RJ, Hattori M, Honda K. 2017. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 358:359–365. doi: 10.1126/science.aan4526. - DOI - PMC - PubMed
-
- Schmidt TS, Hayward MR, Coelho LP, Li SS, Costea PI, Voigt AY, Wirbel J, Maistrenko OM, Alves RJ, Bergsten E, de Beaufort C, Sobhani I, Heintz-Buschart A, Sunagawa S, Zeller G, Wilmes P, Bork P. 2019. Extensive transmission of microbes along the gastrointestinal tract. Elife 8. doi: 10.7554/eLife.42693. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 DK123704/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- F32 AI136404/AI/NIAID NIH HHS/United States
- K01 DK123195/DK/NIDDK NIH HHS/United States
- K01 DK121869/DK/NIDDK NIH HHS/United States
- P30 DK056338/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- F30 DK112563/DK/NIDDK NIH HHS/United States
- T32 DK007664/DK/NIDDK NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- R01 AI123278/AI/NIAID NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- R01 DK103759/DK/NIDDK NIH HHS/United States
- R01 DK115507/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

