High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County
- PMID: 33653938
- PMCID: PMC8546963
- DOI: 10.1128/mSystems.00045-21
High-Throughput Wastewater SARS-CoV-2 Detection Enables Forecasting of Community Infection Dynamics in San Diego County
Abstract
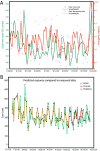
Large-scale wastewater surveillance has the ability to greatly augment the tracking of infection dynamics especially in communities where the prevalence rates far exceed the testing capacity. However, current methods for viral detection in wastewater are severely lacking in terms of scaling up for high throughput. In the present study, we employed an automated magnetic-bead-based concentration approach for viral detection in sewage that can effectively be scaled up for processing 24 samples in a single 40-min run. The method compared favorably to conventionally used methods for viral wastewater concentrations with higher recovery efficiencies from input sample volumes as low as 10 ml and can enable the processing of over 100 wastewater samples in a day. The sensitivity of the high-throughput protocol was shown to detect 1 asymptomatic individual in a building of 415 residents. Using the high-throughput pipeline, samples from the influent stream of the primary wastewater treatment plant of San Diego County (serving 2.3 million residents) were processed for a period of 13 weeks. Wastewater estimates of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral genome copies in raw untreated wastewater correlated strongly with clinically reported cases by the county, and when used alongside past reported case numbers and temporal information in an autoregressive integrated moving average (ARIMA) model enabled prediction of new reported cases up to 3 weeks in advance. Taken together, the results show that the high-throughput surveillance could greatly ameliorate comprehensive community prevalence assessments by providing robust, rapid estimates.IMPORTANCE Wastewater monitoring has a lot of potential for revealing coronavirus disease 2019 (COVID-19) outbreaks before they happen because the virus is found in the wastewater before people have clinical symptoms. However, application of wastewater-based surveillance has been limited by long processing times specifically at the concentration step. Here we introduce a much faster method of processing the samples and show its robustness by demonstrating direct comparisons with existing methods and showing that we can predict cases in San Diego by a week with excellent accuracy, and 3 weeks with fair accuracy, using city sewage. The automated viral concentration method will greatly alleviate the major bottleneck in wastewater processing by reducing the turnaround time during epidemics.
Keywords: COVID-19; SARS-CoV-2; epidemiology; wastewater.
Copyright © 2021 Karthikeyan et al.
Figures
References
-
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wang M, Warren JL, Weinberger DM, Arnold W, Omer SB. 2020. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol 38:1164–1167. doi: 10.1038/s41587-020-0684-z. - DOI - PMC - PubMed
-
- Wu F, Xiao A, Zhang J, Moniz K, Endo N, Armas F, Bonneau R, Brown MA, Bushman M, Chai PR, Duvallet C, Erickson TB, Foppe K, Ghaeli N, Gu X, Hanage WP, Huang KH, Lee WL, Matus M, McElroy KA, Nagler J, Rhode SF, Santillana M, Tucker JA, Wuertz S, Zhao S, Thompson J, Alm EJ. 2020. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. medRxiv 10.1101/2020.06.15.20117747. - DOI - PMC - PubMed
-
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. 2020. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
