Adaptation of pancreatic cancer cells to nutrient deprivation is reversible and requires glutamine synthetase stabilization by mTORC1
- PMID: 33653947
- PMCID: PMC7958225
- DOI: 10.1073/pnas.2003014118
Adaptation of pancreatic cancer cells to nutrient deprivation is reversible and requires glutamine synthetase stabilization by mTORC1
Abstract
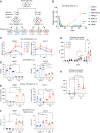
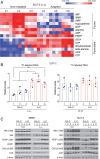
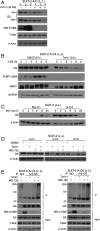
Pancreatic ductal adenocarcinoma (PDA) is a lethal, therapy-resistant cancer that thrives in a highly desmoplastic, nutrient-deprived microenvironment. Several studies investigated the effects of depriving PDA of either glucose or glutamine alone. However, the consequences on PDA growth and metabolism of limiting both preferred nutrients have remained largely unknown. Here, we report the selection for clonal human PDA cells that survive and adapt to limiting levels of both glucose and glutamine. We find that adapted clones exhibit increased growth in vitro and enhanced tumor-forming capacity in vivo. Mechanistically, adapted clones share common transcriptional and metabolic programs, including amino acid use for de novo glutamine and nucleotide synthesis. They also display enhanced mTORC1 activity that prevents the proteasomal degradation of glutamine synthetase (GS), the rate-limiting enzyme for glutamine synthesis. This phenotype is notably reversible, with PDA cells acquiring alterations in open chromatin upon adaptation. Silencing of GS suppresses the enhanced growth of adapted cells and mitigates tumor growth. These findings identify nongenetic adaptations to nutrient deprivation in PDA and highlight GS as a dependency that could be targeted therapeutically in pancreatic cancer patients.
Keywords: epigenetics; glutamine synthetase; mTORC1; nutrient deprivation; pancreatic cancer.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Metabolic plasticity allows cancer cells to thrive under nutrient starvation.Proc Natl Acad Sci U S A. 2021 Apr 5;118(14):e2102057118. doi: 10.1073/pnas.2102057118. Proc Natl Acad Sci U S A. 2021. PMID: 33722932 Free PMC article. No abstract available.
References
-
- Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). - PubMed
-
- Rahib L., et al. ., Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74, 2913–2921 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

