Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins
- PMID: 33654077
- PMCID: PMC7925527
- DOI: 10.1038/s41467-021-21559-9
Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins
Abstract
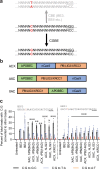
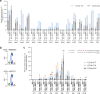
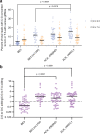
Many genetic diseases are caused by single-nucleotide polymorphisms. Base editors can correct these mutations at single-nucleotide resolution, but until recently, only allowed for transition edits, addressing four out of twelve possible DNA base substitutions. Here, we develop a class of C:G to G:C Base Editors to create single-base genomic transversions in human cells. Our C:G to G:C Base Editors consist of a nickase-Cas9 fused to a cytidine deaminase and base excision repair proteins. Characterization of >30 base editor candidates reveal that they predominantly perform C:G to G:C editing (up to 90% purity), with rAPOBEC-nCas9-rXRCC1 being the most efficient (mean 15.4% and up to 37% without selection). C:G to G:C Base Editors target cytidine in WCW, ACC or GCT sequence contexts and within a precise three-nucleotide window of the target protospacer. We further target genes linked to dyslipidemia, hypertrophic cardiomyopathy, and deafness, showing the therapeutic potential of these base editors in interrogating and correcting human genetic diseases.
Conflict of interest statement
L.C. and W.L.C. are named inventors on a patent application based on this work. Patent applicant: Agency for Science, Technology and Research. Application number: 10201913340Q. Status of Application: PCT submitted; Specific aspect of manuscript covered in patent application: design, applications, and uses of base editors with base excision repair proteins (Figs. 1a, 1b, 3, SI Fig. 3, 5, 7, 9, 10, 11, 13) although all data presented here are included in patent application. The remaining authors declare no competing interests.
Figures
Comment in
-
Base Editing Landscape Extends to Perform Transversion Mutation.Trends Genet. 2020 Dec;36(12):899-901. doi: 10.1016/j.tig.2020.09.001. Epub 2020 Sep 18. Trends Genet. 2020. PMID: 32951947
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

