Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics
- PMID: 33654293
- PMCID: PMC9469728
- DOI: 10.1038/s41591-020-01227-z
Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics
Abstract
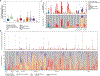
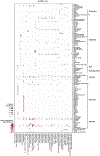
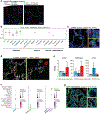
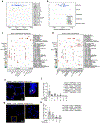
Angiotensin-converting enzyme 2 (ACE2) and accessory proteases (TMPRSS2 and CTSL) are needed for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cellular entry, and their expression may shed light on viral tropism and impact across the body. We assessed the cell-type-specific expression of ACE2, TMPRSS2 and CTSL across 107 single-cell RNA-sequencing studies from different tissues. ACE2, TMPRSS2 and CTSL are coexpressed in specific subsets of respiratory epithelial cells in the nasal passages, airways and alveoli, and in cells from other organs associated with coronavirus disease 2019 (COVID-19) transmission or pathology. We performed a meta-analysis of 31 lung single-cell RNA-sequencing studies with 1,320,896 cells from 377 nasal, airway and lung parenchyma samples from 228 individuals. This revealed cell-type-specific associations of age, sex and smoking with expression levels of ACE2, TMPRSS2 and CTSL. Expression of entry factors increased with age and in males, including in airway secretory cells and alveolar type 2 cells. Expression programs shared by ACE2+TMPRSS2+ cells in nasal, lung and gut tissues included genes that may mediate viral entry, key immune functions and epithelial-macrophage cross-talk, such as genes involved in the interleukin-6, interleukin-1, tumor necrosis factor and complement pathways. Cell-type-specific expression patterns may contribute to the pathogenesis of COVID-19, and our work highlights putative molecular pathways for therapeutic intervention.
Conflict of interest statement
Conflict of interest statement
N.K. was a consultant to Biogen Idec, Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Indaloo, Theravance, LifeMax, Three Lake Partners, Optikira and received non-financial support from MiRagen. All of these outside the work reported. J.L. is a scientific consultant for 10X Genomics Inc. A.R. is a co-founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas, and an SAB member of ThermoFisher Scientific, Syros Pharmaceuticals, Asimov, and Neogene Therapeutics. O.R.R. and A.R. are co-inventors on patent applications filed by the Broad Institute to inventions relating to single cell genomics applications, such as in PCT/US2018/060860 and US Provisional Application No. 62/745,259. A.K.S. compensation for consulting and SAB membership from Honeycomb Biotechnologies, Cellarity, Cogen Therapeutics, Orche Bio, and Dahlia Biosciences. S.A.T. was a consultant at Genentech, Biogen and Roche in the last three years. F.J.T. reports receiving consulting fees from Roche Diagnostics GmbH, and ownership interest in Cellarity Inc. L.V. is founder of Definigen and Bilitech two biotech companies using hPSCs and organoid for disease modelling and cell based therapy. J.A.K. has received advisory board fees from Boehringer Ingelheim, Inc, and has research contracts with Genentech. Eric S. Lander serves on the Board of Directors for Codiak BioSciences and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several non-profit organizations including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. He has served and continues to serve on various federal advisory committees. Joakim Lundeberg is a scientific consultant for 10X Genomics Inc. Jennifer Beane, Joshua Campbell, Mary Reid and Sarah Mazzilli are funded in part by a sponsored research agreement from Janssen Pharmaceuticals, Inc. Avrum Spira is an employee of Johnson & Johnson. Ramnik J. Xavier is a co-founder Celsius Therapeutics and Jnana Therapeutics, and a consultant at Novartis. All other authors declare no conflicts of interest.
Figures











References
Methods references
-
- Seabold S & Perktold J Statsmodels: Econometric and statistical modeling with python. in Proceedings of the 9th Python in Science Conference vol. 57 61 (Austin, TX, 2010).
-
- West BT, Welch KB & Galecki AT Linear Mixed Models: A Practical Guide Using Statistical Software, Second Edition. (CRC Press, 2014).
Publication types
MeSH terms
Substances
Grants and funding
- K08 HL130595/HL/NHLBI NIH HHS/United States
- MR/S005579/1/MRC_/Medical Research Council/United Kingdom
- U01 HL148867/HL/NHLBI NIH HHS/United States
- MR/S035826/1/MRC_/Medical Research Council/United Kingdom
- MR/S035907/1/MRC_/Medical Research Council/United Kingdom
- R01 HL146519/HL/NHLBI NIH HHS/United States
- R01 HL146557/HL/NHLBI NIH HHS/United States
- R01 HL130938/HL/NHLBI NIH HHS/United States
- U01 HL145567/HL/NHLBI NIH HHS/United States
- U01 HL145550/HL/NHLBI NIH HHS/United States
- U01 HL148861/HL/NHLBI NIH HHS/United States
- F32 HL149290/HL/NHLBI NIH HHS/United States
- MR/P009581/1/MRC_/Medical Research Council/United Kingdom
- R01 HL141380/HL/NHLBI NIH HHS/United States
- R01 HL119215/HL/NHLBI NIH HHS/United States
- SP/19/1/34461/BHF_/British Heart Foundation/United Kingdom
- R01 HL127349/HL/NHLBI NIH HHS/United States
- U54 HL165443/HL/NHLBI NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U01 HL148856/HL/NHLBI NIH HHS/United States
- RC2 DK114784/DK/NIDDK NIH HHS/United States
- U19 AI116482/AI/NIAID NIH HHS/United States
- R01 HL145372/HL/NHLBI NIH HHS/United States
- U01 HL122700/HL/NHLBI NIH HHS/United States
- P01 AG049665/AG/NIA NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 DK117263/DK/NIDDK NIH HHS/United States
- K08 HL146943/HL/NHLBI NIH HHS/United States
- U19 AI135964/AI/NIAID NIH HHS/United States
- R01 HL080494/HL/NHLBI NIH HHS/United States
- R24 HD000836/HD/NICHD NIH HHS/United States
- R35 HL140039/HL/NHLBI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U24 AI118672/AI/NIAID NIH HHS/United States
- R01 HL133153/HL/NHLBI NIH HHS/United States
- U19 AI142784/AI/NIAID NIH HHS/United States
- R56 HL135124/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

