In silico evaluation of the compounds of the ayurvedic drug, AYUSH-64, for the action against the SARS-CoV-2 main protease
- PMID: 33654345
- PMCID: PMC7906523
- DOI: 10.1016/j.jaim.2021.02.004
In silico evaluation of the compounds of the ayurvedic drug, AYUSH-64, for the action against the SARS-CoV-2 main protease
Abstract
Background: Outbreak of Corona Virus Disease in late 2019 (COVID-19) has become a pandemic global Public health emergency. Since there is no approved anti-viral drug or vaccine declared for the disease and investigating existing drugs against the COVID-19.
Objective: AYUSH-64 is an Ayurvedic formulation, developed and patented by Central Council of Research in Ayurvedic Sciences, India, has been in clinical use as anti-malarial, anti-inflammatory, anti-pyretic drug for few decades. Thus, the present study was undertaken to evaluate AYUSH-64 compounds available in this drug against Severe Acute Respiratory Syndrome-Corona Virus (SARS-CoV-2) Main Protease (Mpro; PDB ID: 6LU7) via in silico techniques.
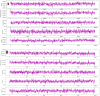
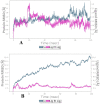
Materials and methods: Different molecular docking software's of Discovery studio and Auto Dock Vina were used for drugs from selected AYUSH-64 compounds against SARS-CoV-2. We also conducted 100 ns period of molecular dynamics simulations with Desmond and further MM/GBSA for the best complex of AYUSH-64 with Mpro of SARS-CoV-2.
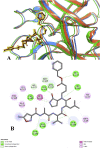
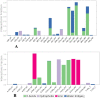
Results: Among 36 compounds of four ingredients of AYUSH-64 screened, 35 observed to exhibits good binding energies than the published positive co-crystal compound of N3 pepetide. The best affinity and interactions of Akuammicine N-Oxide (from Alstonia scholaris) towards the Mpro with binding energy (AutoDock Vina) of -8.4 kcal/mol and Discovery studio of Libdock score of 147.92 kcal/mol. Further, molecular dynamics simulations with MM-GBSA were also performed for Mpro- Akuammicine N-Oxide docked complex to identify the stability, specific interaction between the enzyme and the ligand. Akuammicine N-Oxide is strongly formed h-bonds with crucial Mpro residues, Cys145, and His164.
Conclusion: The results provide lead that, the presence of Mpro- Akuammicine N-Oxide with highest Mpro binding energy along with other 34 chemical compounds having similar activity as part of AYUSH-64 make it a suitable candidate for repurposing to management of COVID-19 by further validating through experimental, clinical studies.
Keywords: AYUSH-64; COVID-19; Dynamics simulations; Main protease; Molecular docking; SARS-CoV-2.
© 2021 The Authors.
Conflict of interest statement
None.
Figures




Similar articles
-
Evaluating therapeutic potential of AYUSH-64 constituents against omicron variant of SARS-CoV-2 using ensemble docking and simulations.Curr Res Struct Biol. 2024 May 31;7:100151. doi: 10.1016/j.crstbi.2024.100151. eCollection 2024. Curr Res Struct Biol. 2024. PMID: 38881558 Free PMC article.
-
Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease.J Biomol Struct Dyn. 2022 Feb;40(2):585-611. doi: 10.1080/07391102.2020.1815584. Epub 2020 Sep 8. J Biomol Struct Dyn. 2022. PMID: 32897178 Free PMC article.
-
In silico validation of coumarin derivatives as potential inhibitors against Main Protease, NSP10/NSP16-Methyltransferase, Phosphatase and Endoribonuclease of SARS CoV-2.J Biomol Struct Dyn. 2021 Nov;39(18):7306-7321. doi: 10.1080/07391102.2020.1808075. Epub 2020 Aug 24. J Biomol Struct Dyn. 2021. PMID: 32835632 Free PMC article.
-
AYUSH- 64: A potential therapeutic agent in COVID-19.J Ayurveda Integr Med. 2022 Apr-Jun;13(2):100538. doi: 10.1016/j.jaim.2021.100538. Epub 2022 Jan 4. J Ayurveda Integr Med. 2022. PMID: 35002178 Free PMC article. Review.
-
Methylxanthines as Potential Inhibitor of SARS-CoV-2: an In Silico Approach.Curr Pharmacol Rep. 2022;8(2):149-170. doi: 10.1007/s40495-021-00276-3. Epub 2022 Mar 8. Curr Pharmacol Rep. 2022. PMID: 35281252 Free PMC article. Review.
Cited by
-
Methodology-Centered Review of Molecular Modeling, Simulation, and Prediction of SARS-CoV-2.Chem Rev. 2022 Jul 13;122(13):11287-11368. doi: 10.1021/acs.chemrev.1c00965. Epub 2022 May 20. Chem Rev. 2022. PMID: 35594413 Free PMC article. Review.
-
Evaluation of Ayush-64 (a Polyherbal Formulation) and Its Ingredients in the Syrian Hamster Model for SARS-CoV-2 Infection Reveals the Preventative Potential of Alstonia scholaris.Pharmaceuticals (Basel). 2023 Sep 21;16(9):1333. doi: 10.3390/ph16091333. Pharmaceuticals (Basel). 2023. PMID: 37765142 Free PMC article.
-
AYUSH-64 as an add-on to standard care in asymptomatic and mild cases of COVID-19: A randomized controlled trial.Ayu. 2020 Apr-Jun;41(2):107-116. doi: 10.4103/ayu.ayu_14_21. Epub 2021 Oct 23. Ayu. 2020. PMID: 34908795 Free PMC article.
-
Protective effects of a small molecule inhibitor ligand against hyperphosphorylated tau-induced mitochondrial and synaptic toxicities in Alzheimer disease.Hum Mol Genet. 2021 Dec 27;31(2):244-261. doi: 10.1093/hmg/ddab244. Hum Mol Genet. 2021. PMID: 34432046 Free PMC article.
-
Plants used in Ayurveda for Jwara or fever: A review of their antiviral studies.J Ayurveda Integr Med. 2025 Mar-Apr;16(2):101085. doi: 10.1016/j.jaim.2024.101085. Epub 2025 Apr 30. J Ayurveda Integr Med. 2025. PMID: 40305981 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

