LINC00504 Promotes the Malignant Biological Behavior of Breast Cancer Cells by Upregulating HMGB3 via Decoying MicroRNA-876-3p
- PMID: 33654429
- PMCID: PMC7910115
- DOI: 10.2147/CMAR.S276290
LINC00504 Promotes the Malignant Biological Behavior of Breast Cancer Cells by Upregulating HMGB3 via Decoying MicroRNA-876-3p
Retraction in
-
LINC00504 Promotes the Malignant Biological Behavior of Breast Cancer Cells by Upregulating HMGB3 via Decoying MicroRNA-876-3p [Retraction].Cancer Manag Res. 2023 Jul 19;15:739-740. doi: 10.2147/CMAR.S430424. eCollection 2023. Cancer Manag Res. 2023. PMID: 37489174 Free PMC article.
Expression of concern in
-
LINC00504 Promotes the Malignant Biological Behavior of Breast Cancer Cells by Upregulating HMGB3 via Decoying MicroRNA-876-3p [Expression of Concern].Cancer Manag Res. 2021 Oct 20;13:7961-7962. doi: 10.2147/CMAR.S344583. eCollection 2021. Cancer Manag Res. 2021. PMID: 34707411 Free PMC article. No abstract available.
Abstract
Purpose: Long intergenic non-protein coding RNA 504 (LINC00504) is a long non-coding RNA that has an important regulatory role in a variety of human cancers. In this study, LINC00504 expression in breast cancer tissues and cell lines was detected. Studies were also conducted to determine the impact of LINC00504 on the tumor behavior of breast cancer cells. The potential mechanisms underlying the oncogenic role of LINC00504 in breast cancer cells were elucidated in detail.
Methods: Expression of LINC00504 in breast cancer was analyzed by quantitative real-time polymerase chain reaction. The effects of LINC00504 on proliferation, apoptosis, in vitro migration and invasion, and in vivo tumor growth were elucidated using Cell Counting Kit-8 assay, flow cytometry, Transwell assays, and tumor xenograft models, respectively. Bioinformatics analyses in conjunction with RNA immunoprecipitation, luciferase reporter assays, and rescue experiments were conducted to investigate the underlying molecular mechanisms.
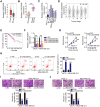
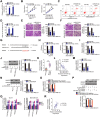
Results: LINC00504 was upregulated in breast cancer tissues and cell lines. Knocking down LINC00504 suppressed breast cancer cell proliferation, migration, and invasion and facilitated apoptosis in vitro. In addition, tumor growth in vivo was significantly inhibited by LINC00504 depletion. Regarding the underlying mechanism, LIN00504 could function as a competing endogenous RNA in breast cancer by sponging microRNA-876-3p (miR-876-3p), resulting in the upregulation of high mobility group box 3 (HMGB3). Rescue experiments further revealed that miR-876-3p downregulation or HMGB3 upregulation effectively reversed the inhibitory effects of LIN00504 deficiency on breast cancer cells.
Conclusion: The LIN00504-miR-876-3p-HMGB3 axis shows carcinogenic effects in modulating the biological behavior of breast cancer cells. This pathway may represent an effective target for CRC diagnosis and anticancer therapy.
Keywords: ceRNA; high mobility group box 3; long intergenic non-protein coding RNA 504; therapeutic target.
© 2021 Yu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

