In-silico nucleotide and protein analyses of S-gene region in selected zoonotic coronaviruses reveal conserved domains and evolutionary emergence with trajectory course of viral entry from SARS-CoV-2 genomic data
- PMID: 33654512
- PMCID: PMC7896521
- DOI: 10.11604/pamj.2020.37.285.24663
In-silico nucleotide and protein analyses of S-gene region in selected zoonotic coronaviruses reveal conserved domains and evolutionary emergence with trajectory course of viral entry from SARS-CoV-2 genomic data
Abstract
Introduction: the recent zoonotic coronavirus virus outbreak of a novel type (COVID-19) has necessitated the adequate understanding of the evolutionary pathway of zoonotic viruses which adversely affects human populations for therapeutic constructs to combat the pandemic now and in the future.
Methods: we analyzed conserved domains of the severe acute respiratory coronavirus 2 (SARS-CoV-2) for possible targets of viral entry inhibition in host cells, evolutionary relationship of human coronavirus (229E) and zoonotic coronaviruses with SARS-CoV-2 as well as evolutionary relationship between selected SARS-CoV-2 genomic data.
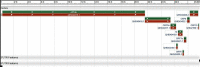
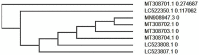
Results: conserved domains with antagonistic action on host innate antiviral cellular mechanisms in SARS-CoV-2 include nsp 11, nsp 13 etc. Also, multiple sequence alignments of the spike (S) gene protein of selected candidate zoonotic coronaviruses alongside the S gene protein of the SARS-CoV-2 revealed closest evolutionary relationship (95.6%) with pangolin coronaviruses (S) gene. Clades formed between Wuhan SARS-CoV-2 phylogeny data and five others suggests viral entry trajectory while revealing genomic and protein SARS-CoV-2 data from Philippines as early ancestors.
Conclusion: phylogeny of SARS-CoV-2 genomic data suggests profiling in diverse populations with and without the outbreak alongside migration history and racial background for mutation tracking and dating of viral subtype divergence which is essential for effective management of present and future zoonotic coronavirus outbreaks.
Keywords: SARS-CoV-2; conserved domains; evolution; phylogeny; spike gene; viral entry; zoonotic CoVs.
Copyright: Adejoke Olukayode Obajuluwa et al.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous