Click Chemistry (CuAAC) and Detection of Tagged de novo Synthesized Proteins in Drosophila
- PMID: 33654887
- PMCID: PMC7854109
- DOI: 10.21769/BioProtoc.3142
Click Chemistry (CuAAC) and Detection of Tagged de novo Synthesized Proteins in Drosophila
Abstract
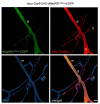
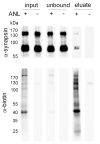
Copper-catalyzed azide-alkyne-cycloaddition (CuAAC), also known as 'click chemistry' serves as a technique for bio-orthogonal, that is, bio-compatible labeling of macromolecules including proteins or lipids. Click chemistry has been widely used to covalently, selectively, and efficiently attach probes such as fluorophores or biotin to small bio-orthogonal chemical reporter groups introduced into macromolecules. In bio-orthogonal non-canonical amino acid tagging (BONCAT) and fluorescent non-canonical amino acid tagging (FUNCAT) proteins are metabolically labeled with a non-canonical, azide-bearing amino acid and subsequently CuAAC-clicked either to an alkyne-bearing biotin (BONCAT) for protein purification, Western blot, or mass spectrometry analyses or to an alkyne-bearing fluorophore (FUNCAT) for immunohistochemistry. In combination with mass spectrometry, these kinds of labeling and tagging strategies are a suitable option to identify and characterize specific proteomes in living organisms without the need of prior cell sorting. Here, we provide detailed protocols for FUNCAT and BONCAT click chemistry and the detection of tagged de novo synthesized proteins in Drosophila melanogaster.
Keywords: Bio-orthogonal chemical reporters; Click chemistry; CuAAC; Drosophila melanogaster; Protein synthesis; Protein tagging; Proteomic profiling.
Copyright © 2019 The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interestsThe authors declare no conflicts of interest or competing interests.
Figures



References
-
- Agard N. J., Prescher J. A. and Bertozzi C. R.(2004). A strain-promoted[3 + 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc 126(46): 15046-15047. - PubMed
-
- Beatty K. E., Liu J. C., Xie F., Dieterich D. C., Schuman E. M., Wang Q. and Tirrell D. A.(2006). Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl 45(44): 7364-7367. - PubMed
-
- Dieterich D. C.(2010). Chemical reporters for the illumination of protein and cell dynamics. Curr Opin Neurobiol 20(5): 623-630. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

