Single-cell mapper (scMappR): using scRNA-seq to infer the cell-type specificities of differentially expressed genes
- PMID: 33655208
- PMCID: PMC7902236
- DOI: 10.1093/nargab/lqab011
Single-cell mapper (scMappR): using scRNA-seq to infer the cell-type specificities of differentially expressed genes
Abstract
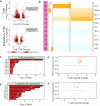
RNA sequencing (RNA-seq) is widely used to identify differentially expressed genes (DEGs) and reveal biological mechanisms underlying complex biological processes. RNA-seq is often performed on heterogeneous samples and the resulting DEGs do not necessarily indicate the cell-types where the differential expression occurred. While single-cell RNA-seq (scRNA-seq) methods solve this problem, technical and cost constraints currently limit its widespread use. Here we present single cell Mapper (scMappR), a method that assigns cell-type specificity scores to DEGs obtained from bulk RNA-seq by leveraging cell-type expression data generated by scRNA-seq and existing deconvolution methods. After evaluating scMappR with simulated RNA-seq data and benchmarking scMappR using RNA-seq data obtained from sorted blood cells, we asked if scMappR could reveal known cell-type specific changes that occur during kidney regeneration. scMappR appropriately assigned DEGs to cell-types involved in kidney regeneration, including a relatively small population of immune cells. While scMappR can work with user-supplied scRNA-seq data, we curated scRNA-seq expression matrices for ∼100 human and mouse tissues to facilitate its stand-alone use with bulk RNA-seq data from these species. Overall, scMappR is a user-friendly R package that complements traditional differential gene expression analysis of bulk RNA-seq data.
© The Author(s) 2021. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

