Development, maturation, and maintenance of human prostate inferred from somatic mutations
- PMID: 33657416
- PMCID: PMC8260206
- DOI: 10.1016/j.stem.2021.02.005
Development, maturation, and maintenance of human prostate inferred from somatic mutations
Abstract
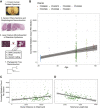
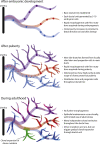
Clonal dynamics and mutation burden in healthy human prostate epithelium are relevant to prostate cancer. We sequenced whole genomes from 409 microdissections of normal prostate epithelium across 8 donors, using phylogenetic reconstruction with spatial mapping in a 59-year-old man's prostate to reconstruct tissue dynamics across the lifespan. Somatic mutations accumulate steadily at ∼16 mutations/year/clone, with higher rates in peripheral than peri-urethral regions. The 24-30 independent glandular subunits are established as rudimentary ductal structures during fetal development by 5-10 embryonic cells each. Puberty induces formation of further side and terminal branches by local stem cells disseminated throughout the rudimentary ducts during development. During adult tissue maintenance, clonal expansions have limited geographic scope and minimal migration. Driver mutations are rare in aging prostate epithelium, but the one driver we did observe generated a sizable intraepithelial clonal expansion. Leveraging unbiased clock-like mutations, we define prostate stem cell dynamics through fetal development, puberty, and aging.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.J.C. is a founder, consultant, and stockholder of Mu Genomics Ltd.
Figures






Comment in
-
A growing genetic tree in the soil of prostate.Cell Stem Cell. 2021 Jul 1;28(7):1185-1187. doi: 10.1016/j.stem.2021.06.002. Cell Stem Cell. 2021. PMID: 34214437
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

