Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India
- PMID: 33658058
- PMCID: PMC7931542
- DOI: 10.1186/s13073-021-00856-4
Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India
Abstract
Background: Type 2 diabetes (T2D), a multifactorial disease influenced by host genetics and environmental factors, is the most common endocrine disease. Several studies have shown that the gut microbiota as a close-up environmental mediator influences host physiology including metabolism. The aim of the present study is to examine the compositional and functional potential of the gut microbiota across individuals from Denmark and South India with a focus on T2D. Many earlier studies have investigated the microbiome aspects of T2D, and it has also been anticipated that such microbial associations would be dependent on diet and ethnic origin. However, there has been no large scale trans-ethnic microbiome study earlier in this direction aimed at evaluating any "universal" microbiome signature of T2D.
Methods: 16S ribosomal RNA gene amplicon sequencing was performed on stool samples from 279 Danish and 294 Indian study participants. Any differences between the gut microbiota of both populations were explored using diversity measures and negative binomial Wald tests. Study samples were stratified to discover global and country-specific microbial signatures for T2D and treatment with the anti-hyperglycemic drug, metformin. To identify taxonomical and functional signatures of the gut microbiota for T2D and metformin treatment, we used alpha and beta diversity measures and differential abundances analysis, comparing metformin-naive T2D patients, metformin-treated T2D patients, and normoglycemic individuals.
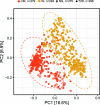
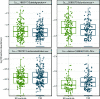
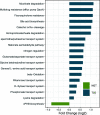
Results: Overall, the gut microbial communities of Danes and Indians are compositionally very different. By analyzing the combined study materials, we identify microbial taxonomic and functional signatures for T2D and metformin treatment. T2D patients have an increased relative abundance of two operational taxonomic units (OTUs) from the Lachnospiraceae family, and a decreased abundance of Subdoligranulum and Butyricicoccus. Studying each population per se, we identified T2D-related microbial changes at the taxonomic level within the Danish population only. Alpha diversity indices show that there is no significant difference between normoglycemic individuals and metformin-naive T2D patients, whereas microbial richness is significantly decreased in metformin-treated T2D patients compared to metformin-naive T2D patients and normoglycemic individuals. Enrichment of two OTUs from Bacteroides and depletion of Faecalibacterium constitute a trans-ethnic signature of metformin treatment.
Conclusions: We demonstrate major compositional differences of the gut microbiota between Danish and South Indian individuals, some of which may relate to differences in ethnicity, lifestyle, and demography. By comparing metformin-naive T2D patients and normoglycemic individuals, we identify T2D-related microbiota changes in the Danish and Indian study samples. In the present trans-ethnic study, we confirm that metformin changes the taxonomic profile and functional potential of the gut microbiota.
Keywords: Danes; Gut microbiota; Indians; Metformin; Populations; Trans-ethnic; Type 2 diabetes.
Conflict of interest statement
SSM, NKP, and AD are affiliated to TCS-Research, Tata Consultancy Services Ltd. (TCS). “TCS-Research” is a DSIR (Govt. of India) recognized R&D unit of TCS, engaging in fundamental as well as translational research activities. Apart from supporting SSM, NKP, and AD with their salaries, TCS, as an organization, had no role to play in the study design, execution, or presentation of results. The remaining authors declare that they have no competing interests.
Figures


References
-
- Cersosimo E, Triplitt C, Solis-Herrera C, Mandarino LJ, DeFronzo RA. Pathogenesis of type 2 diabetes mellitus: MDText.com, Inc.; 2000. http://www.ncbi.nlm.nih.gov/pubmed/25905339. Accessed 12 Mar 2019
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

