Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport
- PMID: 33658209
- PMCID: PMC7929514
- DOI: 10.1126/sciadv.abf5814
Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport
Abstract
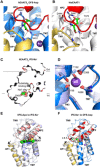
Human excitatory amino acid transporter 3 (hEAAT3) mediates glutamate uptake in neurons, intestine, and kidney. Here, we report cryo-EM structures of hEAAT3 in several functional states where the transporter is empty, bound to coupled sodium ions only, or fully loaded with three sodium ions, a proton, and the substrate aspartate. The structures suggest that hEAAT3 operates by an elevator mechanism involving three functionally independent subunits. When the substrate-binding site is near the cytoplasm, it has a remarkably low affinity for the substrate, perhaps facilitating its release and allowing the rapid transport turnover. The mechanism of the coupled uptake of the sodium ions and the substrate is conserved across evolutionarily distant families and is augmented by coupling to protons in EAATs. The structures further suggest a mechanism by which a conserved glutamate residue mediates proton symport.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Vandenberg R. J., Ryan R. M., Mechanisms of glutamate transport. Physiol. Rev. 93, 1621–1657 (2013). - PubMed
-
- Kanai Y., Hediger M. A., Primary structure and functional characterization of a high-affinity glutamate transporter. Nature 360, 467–471 (1992). - PubMed
-
- Rothstein J. D., Martin L., Levey A. I., Dykes-Hoberg M., Jin L., Wu D., Nash N., Kuncl R. W., Localization of neuronal and glial glutamate transporters. Neuron 13, 713–725 (1994). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

