Interaction of NEP with G Protein Pathway Suppressor 2 Facilitates Influenza A Virus Replication by Weakening the Inhibition of GPS2 to RNA Synthesis and Ribonucleoprotein Assembly
- PMID: 33658351
- PMCID: PMC8139649
- DOI: 10.1128/JVI.00008-21
Interaction of NEP with G Protein Pathway Suppressor 2 Facilitates Influenza A Virus Replication by Weakening the Inhibition of GPS2 to RNA Synthesis and Ribonucleoprotein Assembly
Abstract
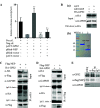
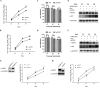
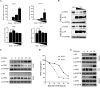
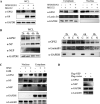
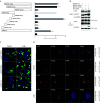
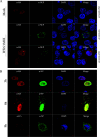
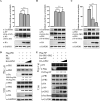
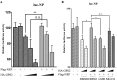
The nuclear export protein (NEP) serves multiple functions in the life cycle of influenza A virus (IAV). Identifying novel host proteins that interact with NEP and understanding their functions in IAV replication are of great interest. In this study, we screened and confirmed the direct interaction of G protein pathway suppressor 2 (GPS2) with NEP through a yeast two-hybrid screening assay and glutathione S-transferase-pulldown and co-immunoprecipitation assays. Knockdown or knockout of GPS2 enhanced IAV titers, whereas overexpression of GPS2 impaired IAV replication, demonstrating that GPS2 acted as a negative host factor in IAV replication. Meanwhile, GPS2 inhibited viral RNA synthesis by reducing the assembly of IAV polymerase. Interestingly, IAV NEP interacted with GPS2 and mediated its nuclear export, thereby activated the degradation of GPS2. Thus, NEP-GPS2 interaction weakened the inhibition of GPS2 to viral polymerase activity and benefited virus replication. Overall, this study identified the novel NEP-binding host partner GPS2 as a critical host factor to participate in IAV replication. These findings provided novel insights into the interactions between IAV and host cells, revealing a new function for GPS2 during IAV replication.Importance: NEP is proposed to play multiple biologically important roles in the life cycle of IAV, which largely relies on host factors by interaction. Our study demonstrated that GPS2 could reduce the interaction between PB1 and PB2 and interfere with vRNP assembly. Thus, GPS2 inhibited the RNA synthesis of IAV and negatively regulated its replication. Importantly, IAV NEP interacted with GPS2 and mediated the nuclear export of GPS2, thereby activated the degradation of GPS2. Thus, NEP-GPS2 interaction weakened the inhibition of GPS2 to viral polymerase activity and benefited virus replication.
Copyright © 2021 Gong et al.
Figures


References
-
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J, Liu W, Zhao G, Yang W, Wang Y, Ma J, Shu Y, Lei F, Gao GF. 2013. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381:1926–1932. doi:10.1016/S0140-6736(13)60938-1. - DOI - PubMed
-
- Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi:10.1038/nature08760. - DOI - PubMed
-
- de Chassey B, Aublin-Gex A, Ruggieri A, Meyniel-Schicklin L, Pradezynski F, Davoust N, Chantier T, Tafforeau L, Mangeot PE, Ciancia C, Perrin-Cocon L, Bartenschlager R, Andre P, Lotteau V. 2013. The interactomes of influenza virus NS1 and NS2 proteins identify new host factors and provide insights for ADAR1 playing a supportive role in virus replication. PLoS Pathog 9:e1003440. doi:10.1371/journal.ppat.1003440. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

