ATF4 selectively regulates heat nociception and contributes to kinesin-mediated TRPM3 trafficking
- PMID: 33658516
- PMCID: PMC7930092
- DOI: 10.1038/s41467-021-21731-1
ATF4 selectively regulates heat nociception and contributes to kinesin-mediated TRPM3 trafficking
Abstract
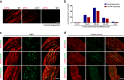
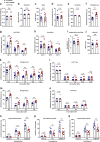
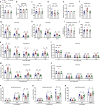
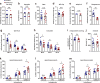
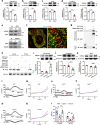
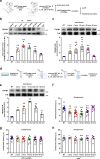
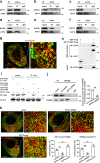
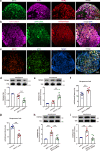
Effective treatments for patients suffering from heat hypersensitivity are lacking, mostly due to our limited understanding of the pathogenic mechanisms underlying this disorder. In the nervous system, activating transcription factor 4 (ATF4) is involved in the regulation of synaptic plasticity and memory formation. Here, we show that ATF4 plays an important role in heat nociception. Indeed, loss of ATF4 in mouse dorsal root ganglion (DRG) neurons selectively impairs heat sensitivity. Mechanistically, we show that ATF4 interacts with transient receptor potential cation channel subfamily M member-3 (TRPM3) and mediates the membrane trafficking of TRPM3 in DRG neurons in response to heat. Loss of ATF4 also significantly decreases the current and KIF17-mediated trafficking of TRPM3, suggesting that the KIF17/ATF4/TRPM3 complex is required for the neuronal response to heat stimuli. Our findings unveil the non-transcriptional role of ATF4 in the response to heat stimuli in DRG neurons.
Conflict of interest statement
All other authors declare no competing interests.
Figures
Similar articles
-
Copine-6 is a TRPM3 escort protein controlling the sensitivity of sensory neurons to noxious heat.EMBO J. 2025 Aug;44(15):4222-4251. doi: 10.1038/s44318-025-00487-0. Epub 2025 Jun 19. EMBO J. 2025. PMID: 40537608 Free PMC article.
-
TRPM3 is a nociceptor channel involved in the detection of noxious heat.Neuron. 2011 May 12;70(3):482-94. doi: 10.1016/j.neuron.2011.02.051. Neuron. 2011. PMID: 21555074
-
Promiscuous G-Protein-Coupled Receptor Inhibition of Transient Receptor Potential Melastatin 3 Ion Channels by Gβγ Subunits.J Neurosci. 2019 Oct 2;39(40):7840-7852. doi: 10.1523/JNEUROSCI.0882-19.2019. Epub 2019 Aug 26. J Neurosci. 2019. PMID: 31451581 Free PMC article.
-
Sensing the heat with TRPM3.Pflugers Arch. 2018 May;470(5):799-807. doi: 10.1007/s00424-017-2100-1. Epub 2018 Jan 5. Pflugers Arch. 2018. PMID: 29305649 Free PMC article. Review.
-
TRPM3 in the eye and in the nervous system - from new findings to novel mechanisms.Biol Chem. 2022 Mar 2;403(8-9):859-868. doi: 10.1515/hsz-2021-0403. Print 2022 Jul 26. Biol Chem. 2022. PMID: 35240732 Review.
Cited by
-
Cochlear organoids reveal transcriptional programs of postnatal hair cell differentiation from supporting cells.Cell Rep. 2023 Nov 28;42(11):113421. doi: 10.1016/j.celrep.2023.113421. Epub 2023 Nov 11. Cell Rep. 2023. PMID: 37952154 Free PMC article.
-
Navigating the Controversies: Role of TRPM Channels in Pain States.Int J Mol Sci. 2024 Sep 24;25(19):10284. doi: 10.3390/ijms251910284. Int J Mol Sci. 2024. PMID: 39408620 Free PMC article. Review.
-
SARS-CoV-2 Airway Infection Results in Time-dependent Sensory Abnormalities in a Hamster Model.bioRxiv [Preprint]. 2022 Aug 19:2022.08.19.504551. doi: 10.1101/2022.08.19.504551. bioRxiv. 2022. Update in: Sci Signal. 2023 May 9;16(784):eade4984. doi: 10.1126/scisignal.ade4984. PMID: 36032984 Free PMC article. Updated. Preprint.
-
Differential immunostaining patterns of transient receptor potential (TRP) ion channels in the rat nodose ganglion.J Anat. 2022 Aug;241(2):230-244. doi: 10.1111/joa.13656. Epub 2022 Apr 9. J Anat. 2022. PMID: 35396708 Free PMC article.
-
A compendium of validated pain genes.WIREs Mech Dis. 2022 Nov;14(6):e1570. doi: 10.1002/wsbm.1570. Epub 2022 Jun 27. WIREs Mech Dis. 2022. PMID: 35760453 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

