Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling
- PMID: 33658518
- PMCID: PMC7930198
- DOI: 10.1038/s41467-021-21615-4
Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling
Abstract
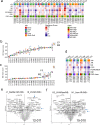
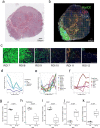
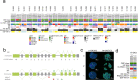
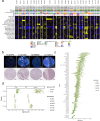
Metastatic prostate cancer (mPC) comprises a spectrum of diverse phenotypes. However, the extent of inter- and intra-tumor heterogeneity is not established. Here we use digital spatial profiling (DSP) technology to quantitate transcript and protein abundance in spatially-distinct regions of mPCs. By assessing multiple discrete areas across multiple metastases, we find a high level of intra-patient homogeneity with respect to tumor phenotype. However, there are notable exceptions including tumors comprised of regions with high and low androgen receptor (AR) and neuroendocrine activity. While the vast majority of metastases examined are devoid of significant inflammatory infiltrates and lack PD1, PD-L1 and CTLA4, the B7-H3/CD276 immune checkpoint protein is highly expressed, particularly in mPCs with high AR activity. Our results demonstrate the utility of DSP for accurately classifying tumor phenotype, assessing tumor heterogeneity, and identifying aspects of tumor biology involving the immunological composition of metastases.
Conflict of interest statement
P.S.N. received instrument support (GeoMx) from NanoString Technologies and consulting fees from Astellas, Janssen, and Bristol Myers Squibb for services unrelated to the present work. M.K., Z.Z., B.B., R.M., G.G., M.H., and J.B. are employees and stockholders of NanoString Technologies. The authors received appropriate permissions for use of NanoString Technologies figures depicting DSP instrument workflow. L.B., C.M., I.C., M.R., L.D.T., R.G., and S.R.P. declare no competing financial interests in relation to this work.
Figures



References
-
- McKenney JK, et al. Histologic grading of prostatic adenocarcinoma can be further optimized: analysis of the relative prognostic strength of individual architectural patterns in 1275 patients from the canary retrospective cohort. Am. J. Surg. Pathol. 2016;40:1439–1456. doi: 10.1097/PAS.0000000000000736. - DOI - PubMed
-
- Clark J, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2006;16:16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

