TRPM2-AS Promotes Bladder Cancer by Targeting miR-22-3p and Regulating GINS2 mRNA Expression
- PMID: 33658791
- PMCID: PMC7914110
- DOI: 10.2147/OTT.S282151
TRPM2-AS Promotes Bladder Cancer by Targeting miR-22-3p and Regulating GINS2 mRNA Expression
Abstract
Background: Bladder cancer (BLCA) refers to the malignancy growth that spreads from the bladder linings to the bladder muscles. However, the impact of miR-22-3p and lncRNA TRPM2-AS on this tumor has generated divergent views in the literature. This research aimed to study the effects of lncRNA TRPM2-AS on BLCA and its interaction with miR-22-3p and GINS2 mRNA.
Methods: qRT-PCR was employed to measure the expression of TRPM2-AS, miR-22-3p and GINS2 mRNA in bladder tissues and cells. The subcellular localization of TRPM2-AS in T24 and 5637 cell lines was identified using a cell fractionation system. Luciferase assay, RIP assay and RNA pull-down assay were later performed to validate the direct binding relationship between TRPM2-AS, miR-22-3p and GINS2 mRNA. Several experiments were conducted to determine the viability, proliferation, colony formation and apoptosis of the cell lines.
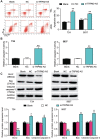
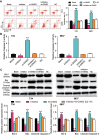
Results: Findings indicated that TRPM2-AS was significantly upregulated in BLCA tissues and cell lines. Apart from that, it was observed that TRPM2-AS knockdown significantly inhibited the viability, proliferation and colony formation of BCLA cells, but it promoted the apoptosis of the BCLA cells. A significant downstream target of TRPM2-AS, miR-22-3p was found to show a lower expression level in BLCA tissues and cell lines. However, the inhibition of miR-22-3p considerably enhanced BLCA cell phenotypes. As well as discovering that GINS2 mRNA was a downstream target of miR-22-3p and was significantly upregulated in BLCA, experimental results also indicated that the knockdown of GINS2 suppressed BLCA cell phenotypes.
Conclusion: This research confirmed that TRPM2-AS could promote BCLA by binding to miR-22-3p to increase GINS2 expression. This novel interactome in BLCA cell lines might provide more insights into BLCA therapy.
Keywords: GINS2; TRPM2-AS; bladder cancer; miR-22-3p.
© 2021 Tian et al.
Conflict of interest statement
There is no conflict of interest existed among the authors.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

