Integrative Analysis of Proteomics and DNA Methylation in Orbital Fibroblasts From Graves' Ophthalmopathy
- PMID: 33658982
- PMCID: PMC7919747
- DOI: 10.3389/fendo.2020.619989
Integrative Analysis of Proteomics and DNA Methylation in Orbital Fibroblasts From Graves' Ophthalmopathy
Abstract
Background: Graves' ophthalmopathy (GO) is a frequent extrathyroidal complication of Graves' hyperthyroidism. Orbital fibroblasts contribute to both orbital tissue inflammation and remodeling in GO, and as such are crucial cellular elements in active GO and inactive GO. However, so far it is largely unknown whether GO disease progression is associated with functional reprogramming of the orbital fibroblast effector function. Therefore, the aim of this study was to compare both the proteome and global DNA methylation patterns between orbital fibroblasts isolated from active GO, inactive GO and healthy controls.
Methods: Orbital fibroblasts from inactive GO (n=5), active GO (n=4) and controls (n=5) were cultured and total protein and DNA was isolated. Labelled and fractionated proteins were analyzed with a liquid chromatography tandem-mass spectrometer (LC-MS/MS). Data are available via ProteomeXchange with identifier PXD022257. Furthermore, bisulphite-treated DNA was analyzed for methylation pattern with the Illumina Infinium Human Methylation 450K beadchip. In addition, RNA was isolated from the orbital fibroblasts for real-time quantitative (RQ)-PCR. Network and pathway analyses were performed.
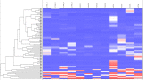
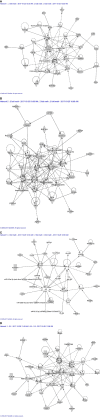
Results: Orbital fibroblasts from active GO displayed overexpression of proteins that are typically involved in inflammation, cellular proliferation, hyaluronan synthesis and adipogenesis, while various proteins associated with extracellular matrix (ECM) biology and fibrotic disease, were typically overexpressed in orbital fibroblasts from inactive GO. Moreover, orbital fibroblasts from active GO displayed hypermethylation of genes that linked to inflammation and hypomethylated genes that linked to adipogenesis and autoimmunity. Further analysis revealed networks that contained molecules to which both hypermethylated and hypomethylated genes were linked, including NF-κB, ERK1/2, Alp, RNA polymerase II, Akt and IFNα. In addition, NF-κB, Akt and IFNα were also identified in networks that were derived from the differentially expressed proteins. Generally, poor correlation between protein expression, DNA methylation and mRNA expression was observed.
Conclusions: Both the proteomics and DNA methylation data support that orbital fibroblasts from active GO are involved in inflammation, adipogenesis, and glycosaminoglycan production, while orbital fibroblasts from inactive disease are more skewed towards an active role in extracellular matrix remodeling. This switch in orbital fibroblast effector function may have therapeutic implications and further studies into the underlying mechanism are thus warranted.
Keywords: DNA methylation; epigenetics; graves’ ophthalmopathy; orbital fibroblast; proteomics.
Copyright © 2021 Virakul, Somparn, Pisitkun, van der Spek, Dalm, Paridaens, van Hagen, Hirankarn, Palaga and Dik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Korfei M, Skwarna S, Henneke I, MacKenzie B, Klymenko O, Saito S, et al. Aberrant expression and activity of histone deacetylases in sporadic idiopathic pulmonary fibrosis. Thorax (2015) 70(11):1022–32. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

