SMART (Single Molecule Analysis of Resection Tracks) Technique for Assessing DNA end-Resection in Response to DNA Damage
- PMID: 33659366
- PMCID: PMC7842315
- DOI: 10.21769/BioProtoc.3701
SMART (Single Molecule Analysis of Resection Tracks) Technique for Assessing DNA end-Resection in Response to DNA Damage
Abstract
DNA double strand breaks (DSBs) are among the most toxic lesions affecting genome integrity. DSBs are mainly repaired through non-homologous end joining (NHEJ) and homologous recombination (HR). A crucial step of the HR process is the generation, through DNA end-resection, of a long 3' single-strand DNA stretch, necessary to prime DNA synthesis using a homologous region as a template, following DNA strand invasion. DNA end resection inhibits NHEJ and triggers homology-directed DSB repair, ultimately guaranteeing a faithful DNA repair. Established methods to evaluate the DNA end-resection process are the immunofluorescence analysis of the phospho-S4/8 RPA32 protein foci, a marker of DNA end-resection, or of the phospho-S4/8 RPA32 protein levels by Western blot. Recently, the Single Molecule Analysis of Resection Tracks (SMART) has been described as a reliable method to visualize, by immunofluorescence, the long 3' single-strand DNA tails generated upon cell treatment with a S-phase specific DNA damaging agent (such as camptothecin). Then, DNA tract lengths can be measured through an image analysis software (such as Photoshop), to evaluate the processivity of the DNA end-resection machinery. The preparation of DNA fibres is performed in non-denaturing conditions so that the immunofluorescence detects only the specific long 3' single-strand DNA tails, generated from DSB processing.
Keywords: BrdU; DNA end-resection; DNA repair; Homologous Recombination; Immunofluorescence.
Copyright © 2020 The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interestsThe authors declare no conflict of interest.
Figures

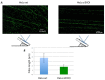

References
-
- Alfano L., Costa C., Caporaso A., Altieri A., Indovina P., Macaluso M., Giordano A. and Pentimalli F.(2016). NONO regulates the intra-S-phase checkpoint in response to UV radiation. Oncogene 35(5): 567-576. - PubMed
-
- Alfano L., Caporaso A., Altieri A., M. Dell'Aquila, Landi C., Bini L., Pentimalli F. and Giordano A.(2019). Depletion of the RNA binding protein HNRNPD impairs homologous recombination by inhibiting DNA-end resection and inducing R-loop accumulation. Nucleic Acids Res 47(8): 4068-4085. - PMC - PubMed
-
- Bianco J. N., Poli J., Saksouk J., Bacal J., Silva M. J., Yoshida K., Lin Y. L., Tourriere H., Lengronne A. and Pasero P.(2012). Analysis of DNA replication profiles in budding yeast and mammalian cells using DNA combing. Methods 57(2): 149-157. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

