Probe-Seq: Method for RNA Sequencing of Specific Cell Types from Animal Tissue
- PMID: 33659409
- PMCID: PMC7842722
- DOI: 10.21769/BioProtoc.3749
Probe-Seq: Method for RNA Sequencing of Specific Cell Types from Animal Tissue
Abstract
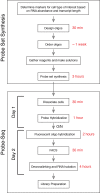
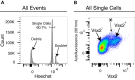
Most organs and tissues are composed of many types of cells. To characterize cellular state, various transcription profiling approaches are currently available, including whole-tissue bulk RNA sequencing, single cell RNA sequencing (scRNA-Seq), and cell type-specific RNA sequencing. What is missing in this repertoire is a simple, versatile method for bulk transcriptional profiling of cell types for which cell type-specific genetic markers or antibodies are not readily available. We therefore developed Probe-Seq, which uses hybridization of gene-specific probes to RNA markers for isolation of specific types of cells, to enable downstream FACS isolation and bulk RNA sequencing. We show that this method can enable isolation and profiling of specific cell types from mouse retina, frozen human retina, Drosophila midgut, and developing chick retina, suggesting that it is likely useful for most organisms.
Keywords: Cell Types; FISH; RNA; RNA-sequencing; Transcriptional Profiling.
Copyright © The Authors; exclusive licensee Bio-protocol LLC.
Conflict of interest statement
Competing interestsThe authors declare no conflict of interest.
Figures

Similar articles
-
Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation.Elife. 2019 Dec 9;8:e51452. doi: 10.7554/eLife.51452. Elife. 2019. PMID: 31815670 Free PMC article.
-
Hydrop enables droplet-based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads.Elife. 2022 Feb 23;11:e73971. doi: 10.7554/eLife.73971. Elife. 2022. PMID: 35195064 Free PMC article.
-
Comparison of cell type distribution between single-cell and single-nucleus RNA sequencing: enrichment of adherent cell types in single-nucleus RNA sequencing.Exp Mol Med. 2022 Dec;54(12):2128-2134. doi: 10.1038/s12276-022-00892-z. Epub 2022 Dec 2. Exp Mol Med. 2022. PMID: 36460793 Free PMC article.
-
Single-cell RNA sequencing: A new opportunity for retinal research.Wiley Interdiscip Rev RNA. 2021 Sep;12(5):e1652. doi: 10.1002/wrna.1652. Epub 2021 Mar 22. Wiley Interdiscip Rev RNA. 2021. PMID: 33754496 Review.
-
Single-cell RNA sequencing in breast cancer: Understanding tumor heterogeneity and paving roads to individualized therapy.Cancer Commun (Lond). 2020 Aug;40(8):329-344. doi: 10.1002/cac2.12078. Epub 2020 Jul 12. Cancer Commun (Lond). 2020. PMID: 32654419 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

