Next-Generation Sequencing in Clinical Practice: Is It a Cost-Saving Alternative to a Single-Gene Testing Approach?
- PMID: 33660227
- PMCID: PMC8160052
- DOI: 10.1007/s41669-020-00249-0
Next-Generation Sequencing in Clinical Practice: Is It a Cost-Saving Alternative to a Single-Gene Testing Approach?
Abstract
Objectives: This study aimed to compare the costs of a next-generation sequencing-based (NGS-based) panel testing strategy to those of a single-gene testing-based (SGT-based) strategy, considering different scenarios of clinical practice evolution.
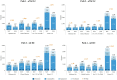
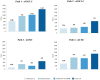
Methods: Three Italian hospitals were analysed, and four different testing pathways (paths 1, 2, 3, and 4) were identified: two for advanced non-small-cell lung cancer (aNSCLC) patients and two for unresectable metastatic colon-rectal cancer (mCRC) patients. For each path, we explored four scenarios considering the current clinical practice and its expected evolution. The 16 testing cases (4 scenarios × 4 paths) were then compared in terms of differential costs between the NGS-based and SGT-based approaches considering personnel, consumables, equipment, and overhead costs. Break-even and sensitivity analyses were performed. Data gathering, aimed at identifying the hospital setup, was performed through a semi-structured questionnaire administered to the professionals involved in testing activities.
Results: The NGS-based strategy was found to be a cost-saving alternative to the SGT-based strategy in 15 of the 16 testing cases. The break-even threshold, the minimum number of patients required to make the NGS-based approach less costly than the SGT-based approach, varied across the testing cases depending on molecular alterations tested, techniques adopted, and specific costs. The analysis found the NGS-based approach to be less costly than the SGT-based approach in nine of the 16 testing cases at any volume of tests performed; in six cases, the NGS-based approach was found to be less costly above a threshold (and in one case, it was found to be always more expensive). Savings obtained using an NGS-based approach ranged from €30 to €1249 per patient; in the unique testing case where NGS was more costly, the additional cost per patient was €25.
Conclusions: An NGS-based approach may be less costly than an SGT-based approach; also, generated savings increase with the number of patients and different molecular alterations tested.
Conflict of interest statement
The authors have no other conflicts of interest to declare.
Figures