Identification of hub lncRNAs in head and neck cancer based on weighted gene co-expression network analysis and experiments
- PMID: 33660438
- PMCID: PMC8406479
- DOI: 10.1002/2211-5463.13134
Identification of hub lncRNAs in head and neck cancer based on weighted gene co-expression network analysis and experiments
Abstract
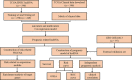
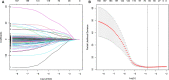
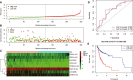
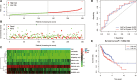
Head and neck squamous cell carcinoma (HNSCC) ranks as the sixth most common cancer among systemic malignant tumors, with 600 000 new cases occurring every year worldwide. Since HNSCC has high heterogeneity and complex pathogenesis, no effective prognostic indicator has yet been identified. Here, we aimed to identify a lncRNA signature associated with the prognosis of HNSCC as a potential new biomarker. LncRNA expression data were downloaded from The Cancer Genome Atlas database. A polygenic risk score model was constructed by using Lasso-Cox regression analysis. Weighted gene co-expression network analysis (WGCNA) was applied to analyze the co-expression modules of lncRNAs associated with the prognosis of HNSCC. The robustness of the signature was validated in testing and external cohorts. Polymerase chain reaction was performed to detect the expression levels of identified lncRNAs in cancer and adjacent tissues. We constructed an 8-lncRNA signature (LINC00567, LINC00996, MTOR-AS1, PRKG1-AS1, RAB11B-AS1, RPS6KA2-AS1, SH3BP5-AS1, ZNF451-AS1) that could be used as an independent prognostic factor of HNSCC. The signature showed strong robustness and had stable prediction performance in different cohorts. WGCNA results showed that modules related to risk score mainly participated in biological processes such as blood vessel development, positive regulation of catabolic processes, and regulation of growth. The prognostic risk score model based on lncRNA for HNSCC may help clinicians conduct individualized treatment plans.
Keywords: The Cancer Genome Atlas; WGCNA; head and neck cancer; risk score prognostic model.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks.Bioengineered. 2021 Dec;12(2):12821-12838. doi: 10.1080/21655979.2021.2003925. Bioengineered. 2021. PMID: 34898376 Free PMC article.
-
Identification and in vitro validation of prognostic lncRNA signature in head and neck squamous cell carcinoma.Bioengineered. 2021 Dec;12(2):10049-10062. doi: 10.1080/21655979.2021.1995577. Bioengineered. 2021. PMID: 34872450 Free PMC article.
-
Constructing a Ferroptosis-related Long Non-coding RNA Signature to Predict the Prognostic of Head and Neck Squamous Cell Carcinoma Patients by Bioinformatic Analysis.Biochem Genet. 2022 Oct;60(5):1825-1844. doi: 10.1007/s10528-021-10176-2. Epub 2022 Jan 31. Biochem Genet. 2022. PMID: 35098409 Free PMC article.
-
Unraveling the oral cancer lncRNAome: Identification of novel lncRNAs associated with malignant progression and HPV infection.Oral Oncol. 2016 Aug;59:58-66. doi: 10.1016/j.oraloncology.2016.05.014. Oral Oncol. 2016. PMID: 27424183 Free PMC article. Review.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
Cited by
-
Identification and Validation of an EMT-Related LncRNA Signature for HNSCC to Predict Survival and Immune Landscapes.Front Cell Dev Biol. 2022 Jan 20;9:798898. doi: 10.3389/fcell.2021.798898. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35273966 Free PMC article.
-
Machine learning-based identification of a novel prognosis-related long noncoding RNA signature for gastric cancer.Front Cell Dev Biol. 2022 Nov 11;10:1017767. doi: 10.3389/fcell.2022.1017767. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36438557 Free PMC article.
-
METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m6A-dependent manner.Cell Mol Biol Lett. 2022 May 20;27(1):41. doi: 10.1186/s11658-022-00342-8. Cell Mol Biol Lett. 2022. PMID: 35596159 Free PMC article.
-
N6-methyladenosine-mediated SH3BP5-AS1 upregulation promotes GEM chemoresistance in pancreatic cancer by activating the Wnt signaling pathway.Biol Direct. 2022 Nov 17;17(1):33. doi: 10.1186/s13062-022-00347-5. Biol Direct. 2022. PMID: 36397058 Free PMC article.
-
Autophagy-related long non-coding RNAs act as prognostic biomarkers and associate with tumor microenvironment in prostate cancer.Am J Cancer Res. 2024 Feb 15;14(2):545-561. doi: 10.62347/XTDZ5687. eCollection 2024. Am J Cancer Res. 2024. PMID: 38455413 Free PMC article.
References
-
- Huang SH and O’Sullivan B (2017) Overview of the 8th edition TNM classification for head and neck cancer. Curr Treat Options Oncol 18, 40. - PubMed
-
- Seeburg DP, Baer AH and Aygun N (2018) Imaging of patients with head and neck cancer: from staging to surveillance. Oral Maxillofac Surg Clin North Am 30, 421–433. - PubMed
-
- Remke M, Radlwimmer B, Muckenthaler M, Breit S, Pfister SJPB (2007) Mutational status of NOTCH1 correlates with specific DNA copy‐number imbalances in acute T‐lineage leukemia. Pediatr Blood Cancer 49, 442–443.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

