Barrier-to-autointegration-factor (Banf1) modulates DNA double-strand break repair pathway choice via regulation of DNA-dependent kinase (DNA-PK) activity
- PMID: 33660778
- PMCID: PMC8034644
- DOI: 10.1093/nar/gkab110
Barrier-to-autointegration-factor (Banf1) modulates DNA double-strand break repair pathway choice via regulation of DNA-dependent kinase (DNA-PK) activity
Abstract
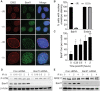
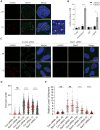
DNA repair pathways are essential to maintain the integrity of the genome and prevent cell death and tumourigenesis. Here, we show that the Barrier-to-Autointegration Factor (Banf1) protein has a role in the repair of DNA double-strand breaks. Banf1 is characterized as a nuclear envelope protein and mutations in Banf1 are associated with the severe premature aging syndrome, Néstor-Guillermo Progeria Syndrome. We have previously shown that Banf1 directly regulates the activity of PARP1 in the repair of oxidative DNA lesions. Here, we show that Banf1 also has a role in modulating DNA double-strand break repair through regulation of the DNA-dependent Protein Kinase catalytic subunit, DNA-PKcs. Specifically, we demonstrate that Banf1 relocalizes from the nuclear envelope to sites of DNA double-strand breaks. We also show that Banf1 can bind to and directly inhibit the activity of DNA-PKcs. Supporting this, cellular depletion of Banf1 leads to an increase in non-homologous end-joining and a decrease in homologous recombination, which our data suggest is likely due to unrestrained DNA-PKcs activity. Overall, this study identifies how Banf1 regulates double-strand break repair pathway choice by modulating DNA-PKcs activity to control genome stability within the cell.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Barrier-to-autointegration factor 1 (Banf1) regulates poly [ADP-ribose] polymerase 1 (PARP1) activity following oxidative DNA damage.Nat Commun. 2019 Dec 3;10(1):5501. doi: 10.1038/s41467-019-13167-5. Nat Commun. 2019. PMID: 31796734 Free PMC article.
-
Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage.BMC Mol Biol. 2012 Mar 9;13:7. doi: 10.1186/1471-2199-13-7. BMC Mol Biol. 2012. PMID: 22404984 Free PMC article.
-
Regulation of the DNA Damage Response by DNA-PKcs Inhibitory Phosphorylation of ATM.Mol Cell. 2017 Jan 5;65(1):91-104. doi: 10.1016/j.molcel.2016.11.004. Epub 2016 Dec 8. Mol Cell. 2017. PMID: 27939942 Free PMC article.
-
DNA-Dependent Protein Kinase Catalytic Subunit (DNA-PKcs): Beyond the DNA Double-Strand Break Repair.Mol Cells. 2023 Apr 30;46(4):200-205. doi: 10.14348/molcells.2023.2164. Epub 2023 Feb 9. Mol Cells. 2023. PMID: 36756777 Free PMC article. Review.
-
Regulation of DNA double-strand break repair pathway choice.Cell Res. 2008 Jan;18(1):134-47. doi: 10.1038/cr.2007.111. Cell Res. 2008. PMID: 18157161 Review.
Cited by
-
The role of inner nuclear membrane proteins in tumourigenesis and as potential targets for cancer therapy.Cancer Metastasis Rev. 2022 Dec;41(4):953-963. doi: 10.1007/s10555-022-10065-z. Epub 2022 Oct 7. Cancer Metastasis Rev. 2022. PMID: 36205821 Free PMC article. Review.
-
Integrated Transcriptomic and Proteomic Study of the Mechanism of Action of the Novel Small-Molecule Positive Allosteric Modulator 1 in Targeting PAC1-R for the Treatment of D-Gal-Induced Aging Mice.Int J Mol Sci. 2024 Mar 30;25(7):3872. doi: 10.3390/ijms25073872. Int J Mol Sci. 2024. PMID: 38612681 Free PMC article.
-
Hutchinson-Gilford progeria patient-derived cardiomyocyte model of carrying LMNA gene variant c.1824 C > T.Cell Tissue Res. 2023 Oct;394(1):189-207. doi: 10.1007/s00441-023-03813-2. Epub 2023 Aug 12. Cell Tissue Res. 2023. PMID: 37572165 Free PMC article.
-
The fructose-bisphosphate, Aldolase A (ALDOA), facilitates DNA-PKcs and ATM kinase activity to regulate DNA double-strand break repair.Sci Rep. 2023 Sep 13;13(1):15171. doi: 10.1038/s41598-023-41133-1. Sci Rep. 2023. PMID: 37704669 Free PMC article.
-
Crosstalk between mitotic reassembly and repair of the nuclear envelope.Nucleus. 2024 Dec;15(1):2352203. doi: 10.1080/19491034.2024.2352203. Epub 2024 May 23. Nucleus. 2024. PMID: 38780365 Free PMC article. Review.
References
-
- Xu Y., Xu D.. Repair pathway choice for double-strand breaks. Essays Biochem. 2020; 64:765–777. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

