The pyruvate:ferredoxin oxidoreductase of the thermophilic acetogen, Thermoanaerobacter kivui
- PMID: 33660937
- PMCID: PMC8091585
- DOI: 10.1002/2211-5463.13136
The pyruvate:ferredoxin oxidoreductase of the thermophilic acetogen, Thermoanaerobacter kivui
Abstract
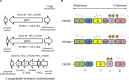
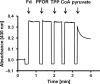
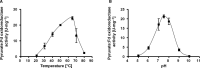
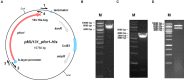
Pyruvate:ferredoxin oxidoreductase (PFOR) is a key enzyme in bacterial anaerobic metabolism. Since a low-potential ferredoxin (Fd2- ) is used as electron carrier, PFOR allows for hydrogen evolution during heterotrophic growth as well as pyruvate synthesis during lithoautotrophic growth. The thermophilic acetogenic model bacterium Thermoanaerobacter kivui can use both modes of lifestyle, but the nature of the PFOR in this organism was previously unestablished. Here, we have isolated PFOR to apparent homogeneity from cells grown on glucose. Peptide mass fingerprinting revealed that it is encoded by pfor1. PFOR uses pyruvate as an electron donor and methylene blue (1.8 U·mg-1 ) and ferredoxin (Fd; 27.2 U·mg-1 ) as electron acceptors, and the reaction is dependent on thiamine pyrophosphate, pyruvate, coenzyme A, and Fd. The pH and temperature optima were 7.5 and 66 °C, respectively. We detected 13.6 mol of iron·mol of protein-1 , consistent with the presence of three predicted [4Fe-4S] clusters. The ability to provide reduced Fd makes PFOR an interesting auxiliary enzyme for enzyme assays. To simplify and speed up the purification procedure, we established a protocol for homologous protein production in T. kivui. Therefore, pfor1 was cloned and expressed in T. kivui and the encoded protein containing a genetically engineered His-tag was purified in only two steps to apparent homogeneity. The homologously produced PFOR1 had the same properties as the enzyme from T. kivui. The enzyme can be used as auxiliary enzyme in enzymatic assays that require reduced Fd as electron donor, such as electron-bifurcating enzymes, to keep a constant level of reduced Fd.
Keywords: extremophile; genetic engineering; homologous gene expression; protein production.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Thauer RK, Kaster AK, Seedorf H, Buckel W and Hedderich R (2008) Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6, 579–591. - PubMed
-
- Drake HL, Gößner AS and Daniel SL (2008) Old acetogens, new light. Ann N Y Acad Sci 1125, 100–128. - PubMed
-
- Schauder R and Kröger A (1993) Bacterial sulphur respiration. Arch Microbiol 159, 491–497.
-
- Ljungdahl LG (1986) The autotrophic pathway of acetate synthesis in acetogenic bacteria. Ann Rev Microbiol 40, 415–450. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

