Activation of skeletal muscle-resident glial cells upon nerve injury
- PMID: 33661767
- PMCID: PMC8119188
- DOI: 10.1172/jci.insight.143469
Activation of skeletal muscle-resident glial cells upon nerve injury
Abstract
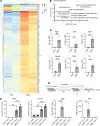
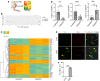
Here, we report on the identification of Itga7-expressing muscle-resident glial cells activated by loss of neuromuscular junction (NMJ) integrity. Gene expression analysis at the bulk and single-cell level revealed that these cells are distinct from Itga7-expressing muscle satellite cells. We show that a selective activation and expansion of Itga7+ glial cells occur in response to muscle nerve lesion. Upon activation, muscle glial-derived progenies expressed neurotrophic genes, including nerve growth factor receptor, which enables their isolation by FACS. We show that activated muscle glial cells also expressed genes potentially implicated in extracellular matrix remodeling at NMJs. We found that tenascin C, which was highly expressed by muscle glial cells, activated upon nerve injury and preferentially localized to NMJ. Interestingly, we observed that the activation of muscle glial cells by acute nerve injury was reversible upon NMJ repair. By contrast, in a mouse model of ALS, in which NMJ degeneration is progressive, muscle glial cells steadily increased over the course of the disease. However, they exhibited an impaired neurotrophic activity, suggesting that pathogenic activation of glial cells may be implicated in ALS progression.
Keywords: Muscle Biology; Neuromuscular disease; Skeletal muscle.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

