Vegetative cells may perform nitrogen fixation function under nitrogen deprivation in Anabaena sp. strain PCC 7120 based on genome-wide differential expression analysis
- PMID: 33662009
- PMCID: PMC7932525
- DOI: 10.1371/journal.pone.0248155
Vegetative cells may perform nitrogen fixation function under nitrogen deprivation in Anabaena sp. strain PCC 7120 based on genome-wide differential expression analysis
Abstract
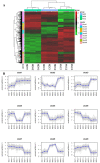
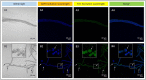
Nitrogen assimilation is strictly regulated in cyanobacteria. In an inorganic nitrogen-deficient environment, some vegetative cells of the cyanobacterium Anabaena differentiate into heterocysts. We assessed the photosynthesis and nitrogen-fixing capacities of heterocysts and vegetative cells, respectively, at the transcriptome level. RNA extracted from nitrogen-replete vegetative cells (NVs), nitrogen-deprived vegetative cells (NDVs), and nitrogen-deprived heterocysts (NDHs) in Anabaena sp. strain PCC 7120 was evaluated by transcriptome sequencing. Paired comparisons of NVs vs. NDHs, NVs vs. NDVs, and NDVs vs. NDHs revealed 2,044 differentially expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the DEGs showed that carbon fixation in photosynthetic organisms and several nitrogen metabolism-related pathways were significantly enriched. Synthesis of Gvp (Gas vesicle synthesis protein gene) in NVs was blocked by nitrogen deprivation, which may cause Anabaena cells to sink and promote nitrogen fixation under anaerobic conditions; in contrast, heterocysts may perform photosynthesis under nitrogen deprivation conditions, whereas the nitrogen fixation capability of vegetative cells was promoted by nitrogen deprivation. Immunofluorescence analysis of nitrogenase iron protein suggested that the nitrogen fixation capability of vegetative cells was promoted by nitrogen deprivation. Our findings provide insight into the molecular mechanisms underlying nitrogen fixation and photosynthesis in vegetative cells and heterocysts at the transcriptome level. This study provides a foundation for further functional verification of heterocyst growth, differentiation, and water bloom control.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
patD, a Gene Regulated by NtcA, Is Involved in the Optimization of Heterocyst Frequency in the Cyanobacterium Anabaena sp. Strain PCC 7120.J Bacteriol. 2019 Oct 4;201(21):e00457-19. doi: 10.1128/JB.00457-19. Print 2019 Nov 1. J Bacteriol. 2019. PMID: 31405917 Free PMC article.
-
Single-Cell Measurements of Fixation and Intercellular Exchange of C and N in the Filaments of the Heterocyst-Forming Cyanobacterium Anabaena sp. Strain PCC 7120.mBio. 2021 Aug 31;12(4):e0131421. doi: 10.1128/mBio.01314-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399619 Free PMC article.
-
Thioredoxin targets are regulated in heterocysts of cyanobacterium Anabaena sp. PCC 7120 in a light-independent manner.J Exp Bot. 2020 Mar 25;71(6):2018-2027. doi: 10.1093/jxb/erz561. J Exp Bot. 2020. PMID: 31863668 Free PMC article.
-
Genetic responses to carbon and nitrogen availability in Anabaena.Environ Microbiol. 2019 Jan;21(1):1-17. doi: 10.1111/1462-2920.14370. Epub 2018 Oct 16. Environ Microbiol. 2019. PMID: 30066380 Review.
-
Cyanobacterial heterocysts.Cold Spring Harb Perspect Biol. 2010 Apr;2(4):a000315. doi: 10.1101/cshperspect.a000315. Epub 2010 Feb 24. Cold Spring Harb Perspect Biol. 2010. PMID: 20452939 Free PMC article. Review.
Cited by
-
Structural basis for glucosylsucrose synthesis by a member of the α-1,2-glucosyltransferase family.Acta Biochim Biophys Sin (Shanghai). 2022 Apr 25;54(4):537-547. doi: 10.3724/abbs.2022034. Acta Biochim Biophys Sin (Shanghai). 2022. PMID: 35607964 Free PMC article.
-
Nitrogen and phosphorus significantly alter growth, nitrogen fixation, anatoxin-a content, and the transcriptome of the bloom-forming cyanobacterium, Dolichospermum.Front Microbiol. 2022 Sep 7;13:955032. doi: 10.3389/fmicb.2022.955032. eCollection 2022. Front Microbiol. 2022. PMID: 36160233 Free PMC article.
References
-
- Rees DC, Kim J, Georgiadis M, Chan MK, Komiya H, Woo D, et al.. 1993. Structures and functions of the nitrogenase proteins, p 83–88. In Palacios R, Mora J, Newton WE (ed), New horizons in nitrogen fixation. Springer, Dordrecht.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

