Polymorphic SERPINA3 prolongs oligomeric state of amyloid beta
- PMID: 33662018
- PMCID: PMC7932536
- DOI: 10.1371/journal.pone.0248027
Polymorphic SERPINA3 prolongs oligomeric state of amyloid beta
Abstract
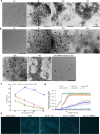
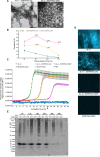
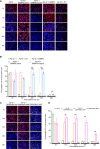
Molecular chaperon SERPINA3 colocalizes with accumulated amyloid peptide in Alzheimer's disease (AD) patient's brain. From the QTL analysis, we narrowed down Serpina3 with two SNPs in senescence-accelerated mouse prone (SAMP) 8 strain. Our study showed SAMP8 type Serpina3 prolonged retention of oligomeric Aβ 42 for longer duration (72 hr) while observing under transmission electron microscope (TEM). From Western blot results, we confirmed presence of Aβ 42 oligomeric forms (trimers, tetramers) were maintained for longer duration only in the presences of SAMP8 type Serpina3. Using SH-SY5Y neuroblastoma cell line, we observed until 36 hr preincubated Aβ 42 with SAMP8 type Serpina3 caused neuronal cell death compared to 12 hr preincubated Aβ 42 with SAMR1 or JF1 type Serpina3 proteins. Similar results were found by extending this study to analyze the effect of polymorphism of SERPINA3 gene of the Japanese SNP database for geriatric research (JG-SNP). We observed that polymorphic SERPINA3 I308T (rs142398813) prolonged toxic oligomeric Aβ 42 forms till 48 hr in comparison to the presence wild type SERPINA3 protein, resulting neuronal cell death. From this study, we first clarified pathogenic regulatory role of polymorphic SERPINA3 in neurodegeneration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

