A comparative analysis of heart microRNAs in vertebrates brings novel insights into the evolution of genetic regulatory networks
- PMID: 33663371
- PMCID: PMC7931589
- DOI: 10.1186/s12864-021-07441-4
A comparative analysis of heart microRNAs in vertebrates brings novel insights into the evolution of genetic regulatory networks
Abstract
Background: During vertebrate evolution, the heart has undergone remarkable changes that lead to morphophysiological differences in the fully formed heart of these species, such as chamber septation, heart rate frequency, blood pressure, and cardiac output volume. Despite these differences, the heart developmental process is guided by a core gene set conserved across vertebrates. Nonetheless, the regulatory mechanisms controlling the expression of genes involved in heart development and maintenance are largely uncharted. MicroRNAs (miRNAs) have been described as important regulatory elements in several biological processes, including heart biology. These small RNA molecules are broadly conserved in sequence and genomic context in metazoans. Mutations may occur in miRNAs and/or genes that contribute to the establishment of distinct repertoires of miRNA-target interactions, thereby favoring the differential control of gene expression and, consequently, the origin of novel phenotypes. In fact, several studies showed that miRNAs are integrated into genetic regulatory networks (GRNs) governing specific developmental programs and diseases. However, studies integrating miRNAs in vertebrate heart GRNs under an evolutionary perspective are still scarce.
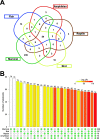
Results: We comprehensively examined and compared the heart miRNome of 20 species representatives of the five major vertebrate groups. We found 54 miRNA families with conserved expression and a variable number of miRNA families with group-specific expression in fishes, amphibians, reptiles, birds, and mammals. We also detected that conserved miRNAs present higher expression levels and a higher number of targets, whereas the group-specific miRNAs present lower expression levels and few targets.
Conclusions: Both the conserved and group-specific miRNAs can be considered modulators orchestrating the core and peripheral genes of heart GRNs of vertebrates, which can be related to the morphophysiological differences and similarities existing in the heart of distinct vertebrate groups. We propose a hypothesis to explain evolutionary differences in the putative functional roles of miRNAs in the heart GRNs analyzed. Furthermore, we present new insights into the molecular mechanisms that could be helping modulate the diversity of morphophysiology in the heart organ of vertebrate species.
Keywords: Cardiac miRNAs; Comparative genomics; Functional genomics; Genetic regulatory network; Non-coding RNA; Small RNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Burggren W, Farrell A, Lillywhite H. Vertebrate cardiovascular systems. Compr Physiol. 2010;:215–308.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

