The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain
- PMID: 33664260
- PMCID: PMC7933334
- DOI: 10.1038/s41467-021-21663-w
The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain
Abstract
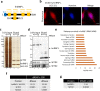
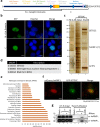
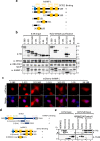
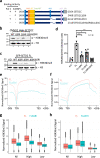
Heterogeneous ribonucleoproteins (hnRNPs) are RNA binding molecules that are involved in key processes such as RNA splicing and transcription. One such hnRNP protein, hnRNP L, regulates alternative splicing (AS) by binding to pre-mRNA transcripts. However, it is unclear what factors contribute to hnRNP L-regulated AS events. Using proteomic approaches, we identified several key factors that co-purify with hnRNP L. We demonstrate that one such factor, the histone methyltransferase SETD2, specifically interacts with hnRNP L in vitro and in vivo. This interaction occurs through a previously uncharacterized domain in SETD2, the SETD2-hnRNP Interaction (SHI) domain, the deletion of which, leads to a reduced H3K36me3 deposition. Functionally, SETD2 regulates a subset of hnRNP L-targeted AS events. Our findings demonstrate that SETD2, by interacting with Pol II as well as hnRNP L, can mediate the crosstalk between the transcription and the splicing machinery.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

