Enzymatic spiroketal formation via oxidative rearrangement of pentangular polyketides
- PMID: 33664266
- PMCID: PMC7933358
- DOI: 10.1038/s41467-021-21432-9
Enzymatic spiroketal formation via oxidative rearrangement of pentangular polyketides
Abstract
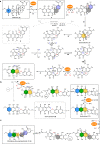
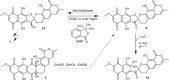
The structural complexity and bioactivity of natural products often depend on enzymatic redox tailoring steps. This is exemplified by the generation of the bisbenzannulated [5,6]-spiroketal pharmacophore in the bacterial rubromycin family of aromatic polyketides, which exhibit a wide array of bioactivities such as the inhibition of HIV reverse transcriptase or DNA helicase. Here we elucidate the complex flavoenzyme-driven formation of the rubromycin pharmacophore that is markedly distinct from conventional (bio)synthetic strategies for spiroketal formation. Accordingly, a polycyclic aromatic precursor undergoes extensive enzymatic oxidative rearrangement catalyzed by two flavoprotein monooxygenases and a flavoprotein oxidase that ultimately results in a drastic distortion of the carbon skeleton. The one-pot in vitro reconstitution of the key enzymatic steps as well as the comprehensive characterization of reactive intermediates allow to unravel the intricate underlying reactions, during which four carbon-carbon bonds are broken and two CO2 become eliminated. This work provides detailed insight into perplexing redox tailoring enzymology that sets the stage for the (chemo)enzymatic production and bioengineering of bioactive spiroketal-containing polyketides.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
An acetyltransferase controls the metabolic flux in rubromycin polyketide biosynthesis by direct modulation of redox tailoring enzymes.Chem Sci. 2022 May 17;13(24):7157-7164. doi: 10.1039/d2sc01952c. eCollection 2022 Jun 22. Chem Sci. 2022. PMID: 35799824 Free PMC article.
-
Catalytic Control of Spiroketal Formation in Rubromycin Polyketide Biosynthesis.Angew Chem Int Ed Engl. 2021 Dec 20;60(52):26960-26970. doi: 10.1002/anie.202109384. Epub 2021 Nov 10. Angew Chem Int Ed Engl. 2021. PMID: 34652045 Free PMC article.
-
Rapid Synthesis of the Spiroketal Subunit of Neaumycin B: Stereochemical Aspects of Singly Anomeric Spiroketals and Proposal for a Stereocenter Reassignment.Org Lett. 2024 Dec 20;26(50):10774-10779. doi: 10.1021/acs.orglett.4c03751. Epub 2024 Dec 9. Org Lett. 2024. PMID: 39652427 Free PMC article.
-
Recent progress in the isolation, bioactivity, biosynthesis, and total synthesis of natural spiroketals.Nat Prod Rep. 2018 Jan 25;35(1):75-104. doi: 10.1039/c7np00043j. Nat Prod Rep. 2018. PMID: 29354841 Review.
-
Benzannulated spiroketal natural products: isolation, biological activity, biosynthesis, and total synthesis.Org Biomol Chem. 2019 Sep 28;17(36):8272-8307. doi: 10.1039/c9ob01598a. Epub 2019 Sep 3. Org Biomol Chem. 2019. PMID: 31478048 Review.
Cited by
-
An acetyltransferase controls the metabolic flux in rubromycin polyketide biosynthesis by direct modulation of redox tailoring enzymes.Chem Sci. 2022 May 17;13(24):7157-7164. doi: 10.1039/d2sc01952c. eCollection 2022 Jun 22. Chem Sci. 2022. PMID: 35799824 Free PMC article.
-
Isolation, biosynthesis, and biological activity of rubromycins derived from actinomycetes.Eng Microbiol. 2022 Jul 31;2(3):100039. doi: 10.1016/j.engmic.2022.100039. eCollection 2022 Sep. Eng Microbiol. 2022. PMID: 39629028 Free PMC article. Review.
-
A Flavoprotein Dioxygenase Steers Bacterial Tropone Biosynthesis via Coenzyme A-Ester Oxygenolysis and Ring Epoxidation.J Am Chem Soc. 2021 Jul 14;143(27):10413-10421. doi: 10.1021/jacs.1c04996. Epub 2021 Jul 1. J Am Chem Soc. 2021. PMID: 34196542 Free PMC article.
-
Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation.Nat Commun. 2022 Sep 14;13(1):5386. doi: 10.1038/s41467-022-33131-0. Nat Commun. 2022. PMID: 36104338 Free PMC article.
-
Three Rings to Rule Them All: How Versatile Flavoenzymes Orchestrate the Structural Diversification of Natural Products.Biochemistry. 2022 Jan 18;61(2):47-56. doi: 10.1021/acs.biochem.1c00763. Epub 2021 Dec 28. Biochemistry. 2022. PMID: 34962769 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

