Site-specific N-glycosylation analysis of animal cell culture-derived Zika virus proteins
- PMID: 33664361
- PMCID: PMC7933209
- DOI: 10.1038/s41598-021-84682-z
Site-specific N-glycosylation analysis of animal cell culture-derived Zika virus proteins
Abstract
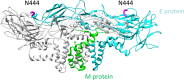
Here, we present for the first time, a site-specific N-glycosylation analysis of proteins from a Brazilian Zika virus (ZIKV) strain. The virus was propagated with high yield in an embryo-derived stem cell line (EB66, Valneva SE), and concentrated by g-force step-gradient centrifugation. Subsequently, the sample was proteolytically digested with different enzymes, measured via a LC-MS/MS-based workflow, and analyzed in a semi-automated way using the in-house developed glyXtoolMS software. The viral non-structural protein 1 (NS1) was glycosylated exclusively with high-mannose structures on both potential N-glycosylation sites. In case of the viral envelope (E) protein, no specific N-glycans could be identified with this method. Nevertheless, N-glycosylation could be proved by enzymatic de-N-glycosylation with PNGase F, resulting in a strong MS-signal of the former glycopeptide with deamidated asparagine at the potential N-glycosylation site N444. This confirmed that this site of the ZIKV E protein is highly N-glycosylated but with very high micro-heterogeneity. Our study clearly demonstrates the progress made towards site-specific N-glycosylation analysis of viral proteins, i.e. for Brazilian ZIKV. It allows to better characterize viral isolates, and to monitor glycosylation of major antigens. The method established can be applied for detailed studies regarding the impact of protein glycosylation on antigenicity and human pathogenicity of many viruses including influenza virus, HIV and corona virus.
Conflict of interest statement
E.R. is the founder and CEO of glyXera GmbH, a company, which offers products and services for glycoanalysis and has several patents in the field. U.R. is a co-owner of the company. All other authors declare no competing interests.
Figures

Similar articles
-
Envelope Protein Glycosylation Mediates Zika Virus Pathogenesis.J Virol. 2019 May 29;93(12):e00113-19. doi: 10.1128/JVI.00113-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944176 Free PMC article.
-
Zika Virus Encoding Nonglycosylated Envelope Protein Is Attenuated and Defective in Neuroinvasion.J Virol. 2017 Nov 14;91(23):e01348-17. doi: 10.1128/JVI.01348-17. Print 2017 Dec 1. J Virol. 2017. PMID: 28931684 Free PMC article.
-
Glycosylation of Zika Virus is Important in Host-Virus Interaction and Pathogenic Potential.Int J Mol Sci. 2019 Oct 21;20(20):5206. doi: 10.3390/ijms20205206. Int J Mol Sci. 2019. PMID: 31640124 Free PMC article.
-
Structures of Zika Virus E & NS1: Relations with Virus Infection and Host Immune Responses.Adv Exp Med Biol. 2018;1062:77-87. doi: 10.1007/978-981-10-8727-1_6. Adv Exp Med Biol. 2018. PMID: 29845526 Review.
-
Advances in Zika Virus⁻Host Cell Interaction: Current Knowledge and Future Perspectives.Int J Mol Sci. 2019 Mar 4;20(5):1101. doi: 10.3390/ijms20051101. Int J Mol Sci. 2019. PMID: 30836648 Free PMC article. Review.
Cited by
-
Glycomics of cervicovaginal fluid from women at risk of preterm birth reveals immuno-regulatory epitopes that are hallmarks of cancer and viral glycosylation.Sci Rep. 2024 Sep 6;14(1):20813. doi: 10.1038/s41598-024-71950-x. Sci Rep. 2024. PMID: 39242814 Free PMC article.
-
Sugar symphony: glycosylation in cancer metabolism and stemness.Trends Cell Biol. 2025 May;35(5):412-425. doi: 10.1016/j.tcb.2024.09.006. Epub 2024 Oct 26. Trends Cell Biol. 2025. PMID: 39462722 Review.
-
Legume Lectins with Different Specificities as Potential Glycan Probes for Pathogenic Enveloped Viruses.Cells. 2022 Jan 20;11(3):339. doi: 10.3390/cells11030339. Cells. 2022. PMID: 35159151 Free PMC article. Review.
-
Dengue and the Lectin Pathway of the Complement System.Viruses. 2021 Jun 24;13(7):1219. doi: 10.3390/v13071219. Viruses. 2021. PMID: 34202570 Free PMC article. Review.
-
Glycolytic inhibitor 2-deoxy-d-glucose attenuates SARS-CoV-2 multiplication in host cells and weakens the infective potential of progeny virions.Life Sci. 2022 Apr 15;295:120411. doi: 10.1016/j.lfs.2022.120411. Epub 2022 Feb 16. Life Sci. 2022. PMID: 35181310 Free PMC article.
References
-
- Del Corral BMS, Rascon JJ, Alonso E. Zika virus associated with microcephaly and abortion, ethical procedures unexplained in relation to an article. Cuad. Bioet. 2016;27:455–457.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

