Polyunsaturated ω-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry
- PMID: 33664446
- PMCID: PMC7933164
- DOI: 10.1038/s41598-021-84850-1
Polyunsaturated ω-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry
Abstract
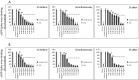
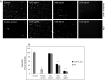
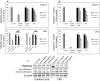
The strain SARS-CoV-2, newly emerged in late 2019, has been identified as the cause of COVID-19 and the pandemic declared by WHO in early 2020. Although lipids have been shown to possess antiviral efficacy, little is currently known about lipid compounds with anti-SARS-CoV-2 binding and entry properties. To address this issue, we screened, overall, 17 polyunsaturated fatty acids, monounsaturated fatty acids and saturated fatty acids, as wells as lipid-soluble vitamins. In performing target-based ligand screening utilizing the RBD-SARS-CoV-2 sequence, we observed that polyunsaturated fatty acids most effectively interfere with binding to hACE2, the receptor for SARS-CoV-2. Using a spike protein pseudo-virus, we also found that linolenic acid and eicosapentaenoic acid significantly block the entry of SARS-CoV-2. In addition, eicosapentaenoic acid showed higher efficacy than linolenic acid in reducing activity of TMPRSS2 and cathepsin L proteases, but neither of the fatty acids affected their expression at the protein level. Also, neither reduction of hACE2 activity nor binding to the hACE2 receptor upon treatment with these two fatty acids was observed. Although further in vivo experiments are warranted to validate the current findings, our study provides a new insight into the role of lipids as antiviral compounds against the SARS-CoV-2 strain.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- International Committee on Taxonomy of Viruses. Taxonomy history: Severe acute respiratory syndrome-related coronavirus" (html). Retrieved 2019-01-27.
-
- Masters, P. S. & Perlman, S. In Fields Virology, Vol. 1 (eds. D. M. Knipe & P. M Howley) Ch. 28, 825–858 (Lippincott Williams & Wilkins, 2013).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

