BCDIN3D RNA methyltransferase stimulates Aldolase C expression and glycolysis through let-7 microRNA in breast cancer cells
- PMID: 33664453
- PMCID: PMC8026734
- DOI: 10.1038/s41388-021-01702-y
BCDIN3D RNA methyltransferase stimulates Aldolase C expression and glycolysis through let-7 microRNA in breast cancer cells
Abstract
Type II diabetes (T2D) and specific cancers share many risk factors, however, the molecular mechanisms underlying these connections are often not well-understood. BCDIN3D is an RNA modifying enzyme that methylates specific precursor microRNAs and tRNAHis. In addition to breast cancer, BCDIN3D may also be linked to metabolism, as its gene locus is associated with obesity and T2D. In order to uncover metabolic pathways regulated by BCDIN3D in cancer, we performed an unbiased analysis of the metabolome, transcriptome, and proteome of breast cancer cells depleted for BCDIN3D. Intersection of these analyses showed that BCDIN3D-depleted cells have increased levels of Fructose 1,6 Bisphosphate (F1,6-BP), the last six-carbon glycolytic intermediate accompanied by reduced glycolytic capacity. We further show that elevated F1,6-BP is due to downregulation of Aldolase C (ALDOC), an enzyme that cleaves F1,6-BP mainly in the brain, but whose high expression/amplification is associated with poor prognosis in breast cancer. BCDIN3D regulates ALDOC through a non-canonical mechanism involving the crucial let-7 microRNA family and its target site on the 3'UTR of ALDOC. Overall, our results reveal an important connection between BCDIN3D, let-7 and glycolysis that may be relevant to breast cancer, obesity, and T2D.
Conflict of interest statement
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
Figures
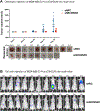

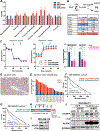
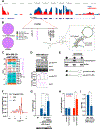
References
-
- Zhu W, Hanes SD. Identification of drosophila bicoid-interacting proteins using a custom two-hybrid selection. Gene 2000; 245: 329–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

