Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy
- PMID: 33664479
- PMCID: PMC8329214
- DOI: 10.1038/s41418-021-00756-5
Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy
Abstract
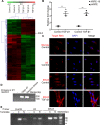
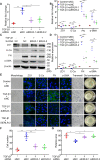
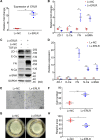
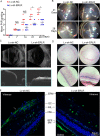
Proliferative vitreoretinopathy (PVR) is a disease that causes severe blindness and is characterized by the formation of contractile fibrotic subretinal or epiretinal membranes. The epithelial-mesenchymal transition (EMT) of retinal pigment epithelial (RPE) cells is a hallmark of PVR. This work aims to examine the role of a long noncoding RNA (lncRNA) named EMT-related lncRNA in RPE (ERLR, LINC01705-201 (ENST00000438158.1)) in PVR and to explore the underlying mechanisms. In this study, we found that ERLR is upregulated in RPE cells stimulated with transforming growth factor (TGF)-β1 as detected by lncRNA microarray and RT-PCR. Further studies characterized full-length ERLR and confirmed that it is mainly expressed in the cytoplasm. In vitro, silencing ERLR in RPE cells attenuated TGF-β1-induced EMT, whereas overexpressing ERLR directly triggered EMT in RPE cells. In vivo, inhibiting ERLR in RPE cells reduced the ability of cells to induce experimental PVR. Mechanistically, chromatin immunoprecipitation (ChIP) assays indicated that the transcription factor TCF4 directly binds to the promoter region of ERLR and promotes its transcription. ERLR mediates EMT by directly binding to MYH9 protein and increasing its stability. TCF4 and MYH9 also mediate TGF-β1-induced EMT in RPE cells. Furthermore, ERLR is also significantly increased in RPE cells incubated with vitreous PVR samples. In clinical samples of PVR membranes, ERLR was detected through fluorescent in situ hybridization (FISH) and colocalized with the RPE marker pancytokeratin (pan-CK). These results indicated that lncRNA ERLR is involved in TGF-β1-induced EMT of human RPE cells and that it is involved in PVR. This finding provides new insights into the mechanism and treatment of PVR.
© 2021. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Long Non-Coding RNA MALAT1 Mediates Transforming Growth Factor Beta1-Induced Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells.PLoS One. 2016 Mar 28;11(3):e0152687. doi: 10.1371/journal.pone.0152687. eCollection 2016. PLoS One. 2016. PMID: 27019196 Free PMC article.
-
Role of semaphorin7A in epithelial-mesenchymal transition and proliferative vitreoretinopathy.Exp Eye Res. 2025 Jan;250:110153. doi: 10.1016/j.exer.2024.110153. Epub 2024 Nov 18. Exp Eye Res. 2025. PMID: 39566570
-
Protective Effects of Fucoidan on Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells and Progression of Proliferative Vitreoretinopathy.Cell Physiol Biochem. 2018;46(4):1704-1715. doi: 10.1159/000489246. Epub 2018 Apr 23. Cell Physiol Biochem. 2018. PMID: 29698960
-
Blockade of Jagged/Notch pathway abrogates transforming growth factor β2-induced epithelial-mesenchymal transition in human retinal pigment epithelium cells.Curr Mol Med. 2014 May;14(4):523-34. doi: 10.2174/1566524014666140331230411. Curr Mol Med. 2014. PMID: 24694299 Review.
-
Mechanisms of epithelial-mesenchymal transition in proliferative vitreoretinopathy.Discov Med. 2015 Oct;20(110):207-17. Discov Med. 2015. PMID: 26562474 Review.
Cited by
-
Prognostication of Pancreatic Cancer Using The Cancer Genome Atlas Based Ferroptosis-Related Long Non-Coding RNAs.Front Genet. 2022 Feb 14;13:838021. doi: 10.3389/fgene.2022.838021. eCollection 2022. Front Genet. 2022. PMID: 35237306 Free PMC article.
-
LINC02086 inhibits ferroptosis and promotes malignant phenotypes of pancreatic cancer via miR-342-3p/CA9 axis.Funct Integr Genomics. 2024 Mar 5;24(2):49. doi: 10.1007/s10142-024-01329-8. Funct Integr Genomics. 2024. PMID: 38438595
-
Tacrolimus Modulates TGF-β Signaling-Related Genes and MicroRNAs in Human Retinal Pigment Epithelial Cells Activated by Lipopolysaccharide.Int J Mol Sci. 2025 Jun 4;26(11):5402. doi: 10.3390/ijms26115402. Int J Mol Sci. 2025. PMID: 40508210 Free PMC article.
-
Flexible Organic Photovoltaic-Powered Hydrogel Bioelectronic Dressing With Biomimetic Electrical Stimulation for Healing Infected Diabetic Wounds.Adv Sci (Weinh). 2024 Mar;11(10):e2307746. doi: 10.1002/advs.202307746. Epub 2023 Dec 25. Adv Sci (Weinh). 2024. PMID: 38145346 Free PMC article.
-
Exosomes-based dual drug-loaded nanocarrier for targeted and multiple proliferative vitreoretinopathy therapy.Regen Biomater. 2024 Jun 29;11:rbae081. doi: 10.1093/rb/rbae081. eCollection 2024. Regen Biomater. 2024. PMID: 39040514 Free PMC article.
References
-
- Yang S, Li H, Li M, Wang F. Mechanisms of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Disco Med. 2015;20:207–17. - PubMed
-
- Bastiaans J, van Meurs JC, van Holten-Neelen C, Nagtzaam NM, van Hagen PM, Chambers RC, et al. Thrombin induces epithelial-mesenchymal transition and collagen production by retinal pigment epithelial cells via autocrine PDGF-receptor signaling. Invest Ophthalmol Vis Sci. 2013;54:8306–14. doi: 10.1167/iovs.13-12383. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

