MACF1 promotes osteoblast differentiation by sequestering repressors in cytoplasm
- PMID: 33664480
- PMCID: PMC8257666
- DOI: 10.1038/s41418-021-00744-9
MACF1 promotes osteoblast differentiation by sequestering repressors in cytoplasm
Abstract
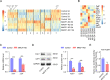
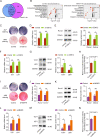
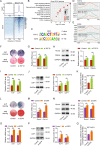
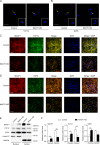
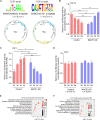
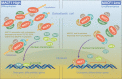
Osteoblast differentiation leading to bone formation requires a coordinated transcriptional program. Osteoblastic cells with low level of microtubule actin crosslinking factor 1 (MACF1) show reduced osteoblast differentiation ability, however, the comprehensive mechanism of MACF1's action remains unexplored. In the current study, we found that MACF1 knockdown suppressed osteoblast differentiation by altering the transcriptome dynamics. We further identified two MACF1-interacted proteins, cyclin-dependent kinase 12 (CDK12) and MYST/Esa1-associated factor 6 (MEAF6), and two MACF1-interacted transcription factors (TFs), transcription factor 12 (TCF12) and E2F transcription factor 6 (E2F6), which repress osteoblast differentiation by altering the expression of osteogenic TFs and genes. Moreover, we found that MACF1 regulated cytoplasmic-nuclear localization of itself, TCF12 and E2F6 in a concentration-dependent manner. MACF1 oppositely regulates the expression of TCF12 and transcription factor 7 (TCF7), two TFs that drive osteoblast differentiation to opposite directions. This study reveals that MACF1, a cytoskeletal protein, acts as a sponge for repressors of osteoblast differentiation to promote osteoblast differentiation and contributes to a novel mechanistic insight of osteoblast differentiation and transcription dynamics.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Microtubule actin crosslinking factor 1 promotes osteoblast differentiation by promoting β-catenin/TCF1/Runx2 signaling axis.J Cell Physiol. 2018 Feb;233(2):1574-1584. doi: 10.1002/jcp.26059. Epub 2017 Aug 3. J Cell Physiol. 2018. PMID: 28621459
-
Mechanical unloading reduces microtubule actin crosslinking factor 1 expression to inhibit β-catenin signaling and osteoblast proliferation.J Cell Physiol. 2018 Jul;233(7):5405-5419. doi: 10.1002/jcp.26374. Epub 2018 Jan 23. J Cell Physiol. 2018. PMID: 29219183
-
MACF1 Overexpression by Transfecting the 21 kbp Large Plasmid PEGFP-C1A-ACF7 Promotes Osteoblast Differentiation and Bone Formation.Hum Gene Ther. 2018 Feb;29(2):259-270. doi: 10.1089/hum.2017.153. Hum Gene Ther. 2018. PMID: 29334773
-
MACF1, versatility in tissue-specific function and in human disease.Semin Cell Dev Biol. 2017 Sep;69:3-8. doi: 10.1016/j.semcdb.2017.05.017. Epub 2017 May 31. Semin Cell Dev Biol. 2017. PMID: 28577926 Review.
-
Isoforms, structures, and functions of versatile spectraplakin MACF1.BMB Rep. 2016 Jan;49(1):37-44. doi: 10.5483/BMBRep.2016.49.1.185. BMB Rep. 2016. PMID: 26521939 Free PMC article. Review.
Cited by
-
Comprehensive ceRNA network for MACF1 regulates osteoblast proliferation.BMC Genomics. 2022 Oct 7;23(1):695. doi: 10.1186/s12864-022-08910-0. BMC Genomics. 2022. PMID: 36207684 Free PMC article.
-
Wnt/β-catenin signaling components and mechanisms in bone formation, homeostasis, and disease.Bone Res. 2024 Jul 10;12(1):39. doi: 10.1038/s41413-024-00342-8. Bone Res. 2024. PMID: 38987555 Free PMC article. Review.
-
Liver cancer stem cell dissemination and metastasis: uncovering the role of NRCAM in hepatocellular carcinoma.J Exp Clin Cancer Res. 2023 Nov 22;42(1):311. doi: 10.1186/s13046-023-02893-w. J Exp Clin Cancer Res. 2023. PMID: 37993901 Free PMC article.
-
The expression profile analysis and functional prediction of lncRNAs in peripheral blood mononuclear cells in maintenance hemodialysis patients developing heart failure.Sci Rep. 2024 Nov 28;14(1):29577. doi: 10.1038/s41598-024-81080-z. Sci Rep. 2024. PMID: 39609580 Free PMC article.
-
MACF1 overexpression in BMSCs alleviates senile osteoporosis in mice through TCF4/miR-335-5p signaling pathway.J Orthop Translat. 2023 Mar 15;39:177-190. doi: 10.1016/j.jot.2023.02.003. eCollection 2023 Mar. J Orthop Translat. 2023. PMID: 36969134 Free PMC article.
References
-
- Ducy P, Schinke T, Karsenty G. The osteoblast: a sophisticated fibroblast under central surveillance. Science. 2000;289:1501–1504. - PubMed
-
- Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. - PubMed
-
- Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. - PubMed
-
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

