Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies
- PMID: 33664494
- PMCID: PMC8058618
- DOI: 10.1038/s41591-021-01294-w
Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global COVID-19 pandemic. Rapidly spreading SARS-CoV-2 variants may jeopardize newly introduced antibody and vaccine countermeasures. Here, using monoclonal antibodies (mAbs), animal immune sera, human convalescent sera and human sera from recipients of the BNT162b2 mRNA vaccine, we report the impact on antibody neutralization of a panel of authentic SARS-CoV-2 variants including a B.1.1.7 isolate, chimeric strains with South African or Brazilian spike genes and isogenic recombinant viral variants. Many highly neutralizing mAbs engaging the receptor-binding domain or N-terminal domain and most convalescent sera and mRNA vaccine-induced immune sera showed reduced inhibitory activity against viruses containing an E484K spike mutation. As antibodies binding to spike receptor-binding domain and N-terminal domain demonstrate diminished neutralization potency in vitro against some emerging variants, updated mAb cocktails targeting highly conserved regions, enhancement of mAb potency or adjustments to the spike sequences of vaccines may be needed to prevent loss of protection in vivo.
Figures









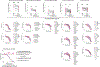




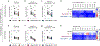
Update of
-
SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies.Res Sq [Preprint]. 2021 Feb 10:rs.3.rs-228079. doi: 10.21203/rs.3.rs-228079/v1. Res Sq. 2021. Update in: Nat Med. 2021 Apr;27(4):717-726. doi: 10.1038/s41591-021-01294-w. PMID: 33594356 Free PMC article. Updated. Preprint.
References
-
- Pinto D, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 75N93019C00074/AI/NIAID NIH HHS/United States
- 75N93019C00062/AI/NIAID NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- R01 AI157155/AI/NIAID NIH HHS/United States
- U01 AI141990/AI/NIAID NIH HHS/United States
- U01 AI151810/AI/NIAID NIH HHS/United States
- U01 AI151801/AI/NIAID NIH HHS/United States
- R01 AI142079/AI/NIAID NIH HHS/United States
- R43 AI145617/AI/NIAID NIH HHS/United States
- UL1 TR001439/TR/NCATS NIH HHS/United States
- 75N93019C00051/AI/NIAID NIH HHS/United States
- R01 AI134907/AI/NIAID NIH HHS/United States
- P30 AR073752/AR/NIAMS NIH HHS/United States
- T32 CA009547/CA/NCI NIH HHS/United States
- HHSN272201400006C/AI/NIAID NIH HHS/United States
- U19 AI142759/AI/NIAID NIH HHS/United States
- F30 AI152327/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

