Wnt-β-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression
- PMID: 33664518
- PMCID: PMC8262575
- DOI: 10.1038/s41590-021-00889-2
Wnt-β-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression
Abstract
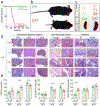
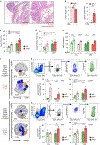
The diversity of regulatory T (Treg) cells in health and in disease remains unclear. Individuals with colorectal cancer harbor a subpopulation of RORγt+ Treg cells with elevated expression of β-catenin and pro-inflammatory properties. Here we show progressive expansion of RORγt+ Treg cells in individuals with inflammatory bowel disease during inflammation and early dysplasia. Activating Wnt-β-catenin signaling in human and murine Treg cells was sufficient to recapitulate the disease-associated increase in the frequency of RORγt+ Treg cells coexpressing multiple pro-inflammatory cytokines. Binding of the β-catenin interacting partner, TCF-1, to DNA overlapped with Foxp3 binding at enhancer sites of pro-inflammatory pathway genes. Sustained Wnt-β-catenin activation induced newly accessible chromatin sites in these genes and upregulated their expression. These findings indicate that TCF-1 and Foxp3 together limit the expression of pro-inflammatory genes in Treg cells. Activation of β-catenin signaling interferes with this function and promotes the disease-associated RORγt+ Treg phenotype.
Figures




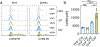










Comment in
-
Wnt-β-catenin as an epigenetic switcher in colonic Treg cells.Nat Immunol. 2021 Apr;22(4):400-401. doi: 10.1038/s41590-021-00904-6. Nat Immunol. 2021. PMID: 33753943 No abstract available.
References
-
- Ito M et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

