Multiple Regionalized Genes and Their Putative Networks in the Interpeduncular Nucleus Suggest Complex Mechanisms of Neuron Development and Axon Guidance
- PMID: 33664652
- PMCID: PMC7921722
- DOI: 10.3389/fnana.2021.643320
Multiple Regionalized Genes and Their Putative Networks in the Interpeduncular Nucleus Suggest Complex Mechanisms of Neuron Development and Axon Guidance
Abstract
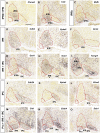
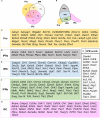
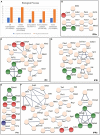
The interpeduncular nucleus (IPN) is a highly conserved limbic structure in the vertebrate brain, located in the isthmus and rhombomere 1. It is formed by various populations that migrate from different sites to the distinct domains within the IPN: the prodromal, rostral interpeduncular, and caudal interpeduncular nuclei. The aim here was to identify genes that are differentially expressed across these domains, characterizing their putative functional roles and interactions. To this end, we screened the 2,038 genes in the Allen Developing Mouse Brain Atlas database expressed at E18.5 and we identified 135 genes expressed within the IPN. The functional analysis of these genes highlighted an overrepresentation of gene families related to neuron development, cell morphogenesis and axon guidance. The interactome analysis within each IPN domain yielded specific networks that mainly involve members of the ephrin/Eph and Cadherin families, transcription factors and molecules related to synaptic neurotransmission. These results bring to light specific mechanisms that might participate in the formation, molecular regionalization, axon guidance and connectivity of the different IPN domains. This genoarchitectonic model of the IPN enables data on gene expression and interactions to be integrated and interpreted, providing a basis for the further study of the connectivity and function of this poorly understood nuclear complex under both normal and pathological conditions.
Keywords: axon guidance; cadherin; ephrin; interactome; interpeduncular nucleus; neuronal migration; rostral hindbrain; transcription factors.
Copyright © 2021 García-Guillén, Alonso, Puelles, Marín and Aroca.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Netrin-1/DCC Signaling Differentially Regulates the Migration of Pax7, Nkx6.1, Irx2, Otp, and Otx2 Cell Populations in the Developing Interpeduncular Nucleus.Front Cell Dev Biol. 2020 Oct 20;8:588851. doi: 10.3389/fcell.2020.588851. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195252 Free PMC article.
-
Interpeduncular nucleus organization in the rat: cytoarchitecture and histochemical analysis.Brain Res Bull. 1984 Jul;13(1):163-79. doi: 10.1016/0361-9230(84)90018-2. Brain Res Bull. 1984. PMID: 6148133
-
Quantitative electron microscopic evidence for reinnervation in the adult rat interpeduncular nucleus after lesions of the fasciculus retroflexus.J Comp Neurol. 1979 Sep 15;187(2):447-68. doi: 10.1002/cne.901870211. J Comp Neurol. 1979. PMID: 489788
-
Hox genes in neural patterning and circuit formation in the mouse hindbrain.Curr Top Dev Biol. 2009;88:139-67. doi: 10.1016/S0070-2153(09)88005-8. Curr Top Dev Biol. 2009. PMID: 19651304 Review.
-
The habenulo-interpeduncular pathway in nicotine aversion and withdrawal.Neuropharmacology. 2015 Sep;96(Pt B):213-22. doi: 10.1016/j.neuropharm.2014.11.019. Epub 2014 Dec 2. Neuropharmacology. 2015. PMID: 25476971 Free PMC article. Review.
Cited by
-
An atlas of transcriptionally defined cell populations in the rat ventral tegmental area.Cell Rep. 2022 Apr 5;39(1):110616. doi: 10.1016/j.celrep.2022.110616. Cell Rep. 2022. PMID: 35385745 Free PMC article.
-
Prolonged nicotine exposure reduces aversion to the drug in mice by altering nicotinic transmission in the interpeduncular nucleus.Elife. 2023 May 30;12:e80767. doi: 10.7554/eLife.80767. Elife. 2023. PMID: 37249215 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

