Cellular and Genomic Properties of Haloferax gibbonsii LR2-5, the Host of Euryarchaeal Virus HFTV1
- PMID: 33664716
- PMCID: PMC7921747
- DOI: 10.3389/fmicb.2021.625599
Cellular and Genomic Properties of Haloferax gibbonsii LR2-5, the Host of Euryarchaeal Virus HFTV1
Abstract
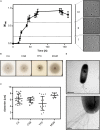
Hypersaline environments are the source of many viruses infecting different species of halophilic euryarchaea. Information on infection mechanisms of archaeal viruses is scarce, due to the lack of genetically accessible virus-host models. Recently, a new archaeal siphovirus, Haloferax tailed virus 1 (HFTV1), was isolated together with its host belonging to the genus Haloferax, but it is not infectious on the widely used model euryarcheon Haloferax volcanii. To gain more insight into the biology of HFTV1 host strain LR2-5, we studied characteristics that might play a role in its virus susceptibility: growth-dependent motility, surface layer, filamentous surface structures, and cell shape. Its genome sequence showed that LR2-5 is a new strain of Haloferax gibbonsii. LR2-5 lacks obvious viral defense systems, such as CRISPR-Cas, and the composition of its cell surface is different from Hfx. volcanii, which might explain the different viral host range. This work provides first deep insights into the relationship between the host of halovirus HFTV1 and other members of the genus Haloferax. Given the close relationship to the genetically accessible Hfx. volcanii, LR2-5 has high potential as a new model for virus-host studies in euryarchaea.
Keywords: N-glycosylation; S-layer; archaeal virus; archaellum; haloarchaea; type IV pili.
Copyright © 2021 Tittes, Schwarzer, Pfeiffer, Dyall-Smith, Rodriguez-Franco, Oksanen and Quax.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The Viral Susceptibility of the Haloferax Species.Viruses. 2022 Jun 20;14(6):1344. doi: 10.3390/v14061344. Viruses. 2022. PMID: 35746816 Free PMC article.
-
Development of a genetic system for Haloferax gibbonsii LR2-5, model host for haloarchaeal viruses.Appl Environ Microbiol. 2024 Apr 17;90(4):e0012924. doi: 10.1128/aem.00129-24. Epub 2024 Mar 12. Appl Environ Microbiol. 2024. PMID: 38470030 Free PMC article.
-
Archaeal Host Cell Recognition and Viral Binding of HFTV1 to Its Haloferax Host.mBio. 2023 Feb 28;14(1):e0183322. doi: 10.1128/mbio.01833-22. Epub 2023 Jan 19. mBio. 2023. PMID: 36656006 Free PMC article.
-
Archaeal membrane-associated proteases: insights on Haloferax volcanii and other haloarchaea.Front Microbiol. 2015 Feb 6;6:39. doi: 10.3389/fmicb.2015.00039. eCollection 2015. Front Microbiol. 2015. PMID: 25774151 Free PMC article. Review.
-
Add salt, add sugar: N-glycosylation in Haloferax volcanii.Biochem Soc Trans. 2013 Feb 1;41(1):432-5. doi: 10.1042/BST20120142. Biochem Soc Trans. 2013. PMID: 23356324 Review.
Cited by
-
Open Issues for Protein Function Assignment in Haloferax volcanii and Other Halophilic Archaea.Genes (Basel). 2021 Jun 24;12(7):963. doi: 10.3390/genes12070963. Genes (Basel). 2021. PMID: 34202810 Free PMC article.
-
"Influence of plasmids, selection markers and auxotrophic mutations on Haloferax volcanii cell shape plasticity".Front Microbiol. 2023 Sep 29;14:1270665. doi: 10.3389/fmicb.2023.1270665. eCollection 2023. Front Microbiol. 2023. PMID: 37840741 Free PMC article.
-
The Viral Susceptibility of the Haloferax Species.Viruses. 2022 Jun 20;14(6):1344. doi: 10.3390/v14061344. Viruses. 2022. PMID: 35746816 Free PMC article.
-
Understanding the tolerance of halophilic archaea to stress landscapes.Environ Microbiol Rep. 2024 Dec;16(6):e70039. doi: 10.1111/1758-2229.70039. Environ Microbiol Rep. 2024. PMID: 39568122 Free PMC article.
-
Growth Phase Dependent Cell Shape of Haloarcula.Microorganisms. 2021 Jan 22;9(2):231. doi: 10.3390/microorganisms9020231. Microorganisms. 2021. PMID: 33499340 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

