Comparison of Two 16S rRNA Primers (V3-V4 and V4-V5) for Studies of Arctic Microbial Communities
- PMID: 33664723
- PMCID: PMC7920977
- DOI: 10.3389/fmicb.2021.637526
Comparison of Two 16S rRNA Primers (V3-V4 and V4-V5) for Studies of Arctic Microbial Communities
Abstract
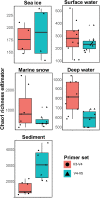
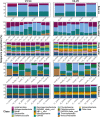
Microbial communities of the Arctic Ocean are poorly characterized in comparison to other aquatic environments as to their horizontal, vertical, and temporal turnover. Yet, recent studies showed that the Arctic marine ecosystem harbors unique microbial community members that are adapted to harsh environmental conditions, such as near-freezing temperatures and extreme seasonality. The gene for the small ribosomal subunit (16S rRNA) is commonly used to study the taxonomic composition of microbial communities in their natural environment. Several primer sets for this marker gene have been extensively tested across various sample sets, but these typically originated from low-latitude environments. An explicit evaluation of primer-set performances in representing the microbial communities of the Arctic Ocean is currently lacking. To select a suitable primer set for studying microbiomes of various Arctic marine habitats (sea ice, surface water, marine snow, deep ocean basin, and deep-sea sediment), we have conducted a performance comparison between two widely used primer sets, targeting different hypervariable regions of the 16S rRNA gene (V3-V4 and V4-V5). We observed that both primer sets were highly similar in representing the total microbial community composition down to genus rank, which was also confirmed independently by subgroup-specific catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) counts. Each primer set revealed higher internal diversity within certain bacterial taxonomic groups (e.g., the class Bacteroidia by V3-V4, and the phylum Planctomycetes by V4-V5). However, the V4-V5 primer set provides concurrent coverage of the archaeal domain, a relevant component comprising 10-20% of the community in Arctic deep waters and the sediment. Although both primer sets perform similarly, we suggest the use of the V4-V5 primer set for the integration of both bacterial and archaeal community dynamics in the Arctic marine environment.
Keywords: Arctic Ocean; amplicon sequencing; method comparison; microbial communities; molecular observatory; universal primers.
Copyright © 2021 Fadeev, Cardozo-Mino, Rapp, Bienhold, Salter, Salman-Carvalho, Molari, Tegetmeyer, Buttigieg and Boetius.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75 129–137. 10.3354/ame01753 - DOI
-
- Armougom F. (2009). Exploring microbial diversity using 16S rRNA high-throughput methods. J. Comput. Sci. Syst. Biol. 2 74–92. 10.4172/jcsb.1000019 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

