PDIA5 is Correlated With Immune Infiltration and Predicts Poor Prognosis in Gliomas
- PMID: 33664747
- PMCID: PMC7921737
- DOI: 10.3389/fimmu.2021.628966
PDIA5 is Correlated With Immune Infiltration and Predicts Poor Prognosis in Gliomas
Abstract
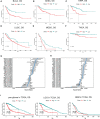
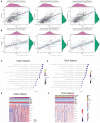
Gliomas are the most common and lethal primary malignant tumor of the brain. Routine treatment including surgical resection, chemotherapy, and radiotherapy produced limited therapeutic effect, while immunotherapy targeting the glioma microenvironment has offered a novel therapeutic option. PDIA5 protein is the member of PDI family, which is highly expressed in glioma and participates in glioma progression. Based on large-scale bioinformatics analysis, we discovered that PDIA5 expression level is upregulated in aggressive gliomas, with high PDIA5 expression predicting poor clinical outcomes. We also observed positive correlation between PDIA5 and immune infiltrating cells, immune related pathways, inflammatory activities, and other immune checkpoint members. Patients with high PDIA5 high-expression benefited from immunotherapies. Additionally, immunohistochemistry revealed that PDIA5 and macrophage biomarker CD68 were upregulated in high-grade gliomas, and patients with low PDIA5 level experienced favorable outcomes among 33 glioma patients. Single cell RNA sequencing exhibited that PDIA5 was in high level presenting in neoplastic cells and macrophages. Cell transfection and co-culture of glioma cells and macrophages revealed that PDIA5 in tumor cells mediated macrophages exhausting. Altogether, our findings indicate that PDIA5 overexpression is associated with immune infiltration in gliomas, and may be a promising therapeutic target for glioma immunotherapy.
Keywords: PDIA5; gliomas; immune infiltration; immunotherapy; scRNA-seq.
Copyright © 2021 Zhang, He, Dai, Wang, Liang, He, Xia, Feng, Cao, Zhang and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Protein disulfide isomerases are promising targets for predicting the survival and tumor progression in glioma patients.Aging (Albany NY). 2020 Feb 5;12(3):2347-2372. doi: 10.18632/aging.102748. Epub 2020 Feb 5. Aging (Albany NY). 2020. PMID: 32023222 Free PMC article.
-
TNFSF13 Is a Novel Onco-Inflammatory Marker and Correlates With Immune Infiltration in Gliomas.Front Immunol. 2021 Oct 12;12:713757. doi: 10.3389/fimmu.2021.713757. eCollection 2021. Front Immunol. 2021. PMID: 34712225 Free PMC article.
-
Orphan nuclear receptor TLX promotes immunosuppression via its transcriptional activation of PD-L1 in glioma.J Immunother Cancer. 2021 Apr;9(4):e001937. doi: 10.1136/jitc-2020-001937. J Immunother Cancer. 2021. PMID: 33858847 Free PMC article.
-
Immunotherapy for gliomas: shedding light on progress in preclinical and clinical development.Expert Opin Investig Drugs. 2020 Jul;29(7):659-684. doi: 10.1080/13543784.2020.1768528. Epub 2020 Jun 4. Expert Opin Investig Drugs. 2020. PMID: 32400216 Review.
-
Dendritic cell immunotherapy for malignant gliomas.Rev Recent Clin Trials. 2008 Jan;3(1):10-21. doi: 10.2174/157488708783330530. Rev Recent Clin Trials. 2008. PMID: 18474011 Review.
Cited by
-
Novel Immune Infiltrating Cell Signature Based on Cell Pair Algorithm Is a Prognostic Marker in Cancer.Front Immunol. 2021 Sep 14;12:694490. doi: 10.3389/fimmu.2021.694490. eCollection 2021. Front Immunol. 2021. PMID: 34594324 Free PMC article.
-
IP-score correlated to endogenous tumour antigen peptide processing: A candidate clinical response score algorithm of immune checkpoint inhibitors therapy in multiple cohorts.Front Immunol. 2023 Jan 9;13:1085491. doi: 10.3389/fimmu.2022.1085491. eCollection 2022. Front Immunol. 2023. PMID: 36700205 Free PMC article.
-
Role of CD68 in tumor immunity and prognosis prediction in pan-cancer.Sci Rep. 2022 May 12;12(1):7844. doi: 10.1038/s41598-022-11503-2. Sci Rep. 2022. PMID: 35550532 Free PMC article.
-
Integrin Alpha-2 as a Potential Prognostic and Predictive Biomarker for Patients With Lower-Grade Glioma.Front Oncol. 2021 Oct 27;11:738651. doi: 10.3389/fonc.2021.738651. eCollection 2021. Front Oncol. 2021. PMID: 34778054 Free PMC article.
-
Nuclear mitochondria-related genes-based molecular classification and prognostic signature reveal immune landscape, somatic mutation, and prognosis for glioma.Heliyon. 2023 Sep 5;9(9):e19856. doi: 10.1016/j.heliyon.2023.e19856. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809472 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

